Abstract
Purpose
CYP2D6 bioactivates codeine and tramadol, with intermediate and poor metabolizers (IMs and PMs) expected to have impaired analgesia. This pragmatic proof-of-concept trial tested the effects of CYP2D6-guided opioid prescribing on pain control.
Methods
Participants with chronic pain (94% on an opioid) from seven clinics were enrolled into CYP2D6-guided (n = 235) or usual care (n = 135) arms using a cluster design. CYP2D6 phenotypes were assigned based on genotype and CYP2D6 inhibitor use, with recommendations for opioid prescribing made in the CYP2D6-guided arm. Pain was assessed at baseline and 3 months using PROMIS® measures.
Results
On stepwise multiple linear regression, the primary outcome of composite pain intensity (composite of current pain and worst and average pain in the past week) among IM/PMs initially prescribed tramadol/codeine (n = 45) had greater improvement in the CYP2D6-guided versus usual care arm (-1.01 ± 1.59 vs. -0.40 ± 1.20; adj P = 0.016); 24% of CYP2D6-guided versus 0% of usual care participants reported ≥30% (clinically meaningful) reduction in the composite outcome. In contrast, among normal metabolizers prescribed tramadol or codeine at baseline, there was no difference in the change in composite pain intensity at 3 months between CYP2D6-guided (-0.61 ± 1.39) and usual care (-0.54 ± 1.69) groups (adj P = 0.540).
Conclusion
These data support the potential benefits of CYP2D6-guided pain management.
Similar content being viewed by others
INTRODUCTION
Over 100 million Americans suffer from pain,1 and opioids are widely prescribed for pain management. Nearly half of all opioid prescriptions are from primary care practice settings.2 Overprescribing of opioids is widely considered to contribute to the opioid crisis of misuse, abuse, and overdose.3 Therefore, it is important to implement primary care interventions that improve assessment of opioid-related risks and benefits and guide safe and effective prescribing for pain.
Tramadol, codeine, hydrocodone, and oxycodone comprise the majority of opioids prescribed in the United States.4 The cytochrome P450 2D6 (CYP2D6) enzyme bioactivates codeine to morphine and tramadol to O-desmethyltramadol (M1), which have 200-fold greater affinity for the µ-opioid receptor than their parent compounds.5 Thus, the parent compounds have little to no analgesic effect relative to their metabolites.6 CYP2D6 also converts oxycodone to oxymorphone and hydrocodone to hydromorphone, which also have higher affinity for the µ-opioid receptor than their parent compounds.5
The CYP2D6 gene is highly polymorphic, which translates to enzymatic activities ranging from nonfunctional (i.e., poor metabolizers; PM) to increased function (i.e., ultrarapid metabolizers; UM). The CYP2D6*3, *4, *5, and *6 alleles are nonfunctional alleles that occur secondary to frameshift variants (*3, *6), splicing defects (*4), or whole-gene deletion (*5).5 PMs have two nonfunctional alleles and cannot metabolize CYP2D6 substrates. Compared with normal metabolizers (NMs), PMs have lower concentrations of the active metabolites of codeine, tramadol, hydrocodone, or oxycodone.7,8,9,10 Intermediate metabolizers (IMs) possess one nonfunctional and one reduced function allele and have significantly reduced enzyme activity compared with NMs.5 Although less well studied, IMs are at risk of reduced biotransformation of CYP2D6-mediated opioids to more potent metabolites, conferring decreased analgesia.5,11 On the other hand, UMs have multiple copies of functional alleles and are at risk for toxic concentrations of active metabolites of CYP2D6-metabolized opioids, with reports of life-threatening toxicity and death.12,13,14,15 The frequencies of PM, IM, and UM phenotypes range from 5% to 10%, 5% to 11%, and 3% to 29%, respectively, in different populations.5,16
Concomitant use of CYP2D6 inhibitors can also reduce CYP2D6-mediated metabolism and cause phenoconversion, which changes the phenotype the patient would have based on their genetics. For example paroxetine, a strong CYP2D6 inhibitor,17 has been shown to phenoconvert patients genotypically classified as NMs to PMs. Phenoconversion is likely common, as data indicate approximately 20–30% of patients treated for pain are prescribed a CYP2D6 inhibitor.18,19
Prescribing of opioids metabolized by CYP2D6 represents an excellent opportunity for precision medicine given common CYP2D6 genotype variability and frequent CYP2D6 inhibitor use, both of which may influence efficacy of opioids metabolized via CYP2D6. The Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines recommend against codeine use in CYP2D6 PMs and UMs and advise against use of tramadol, and to a lesser extent, hydrocodone and oxycodone as alternatives.5 Although the data are less clear, genotyping may also be informative for hydrocodone or oxycodone prescribing. To our knowledge, there are no data on the effect of clinical implementation of a CYP2D6-guided approach to opioid prescribing on clinical outcomes of pain control. This trial tested whether CYP2D6-guided opioid prescribing improves chronic pain control for patients managed in primary care and specialty clinics.
MATERIALS AND METHODS
Design
This pragmatic, proof-of-concept trial employed a nonrandomized, open-label, prospective, cluster design to compare CYP2D6-guided versus usual management of chronic pain over 3 months. According to the PRagmatic-Explanatory Continuum Indicator Summary (PRECIS)-2 tool, this trial was highly pragmatic as opposed to explanatory in seven of nine domains assessed (Table S1).20 Patients were enrolled between May 2015 and June 2017 from seven University of Florida (UF) Health primary care clinics designated as either CYP2D6-guided (n = 4) or usual care (n = 3) clinics.
Study participants
Eligible participants were ≥18 years of age; being managed for chronic pain, defined as pain ≥3 months, at one of the participating clinics; and receiving or eligible to receive an opioid. Patients with prior CYP2D6 genotype results were excluded. There were no inclusion criteria related to timing of opioid initiation, type of opioid used, or level of pain control. Patients were referred for study participation by physicians in participating clinics. Physicians were encouraged to refer patients with poorly controlled pain or patients for whom a change in pain therapy was being considered. All participants provided written, informed consent. The study was approved by the UF Institutional Review Board, and all procedures were in accordance with the ethical standards of the Declaration of Helsinki. The trial is registered at ClinicalTrials.gov: NCT02335307.
Intervention
Participants enrolled from CYP2D6-guided clinics provided a buccal cell sample for CYP2D6 genotyping, with genotype results reported in their electronic health record (EHR). Participants enrolled at usual care clinics also provided a buccal cell sample, which was stored until the end of their participation (3 months) and then used for genotyping if the participant desired, with results reported in their EHR.
CYP2D6 genotyping and phenotype assignment
CYP2D6 genotype was determined using a Luminex xTAG CYP2D6 Kit v3 (Luminex Corporation, Austin, TX, USA) at the UF Health Pathology Laboratories. Each allele was assigned an activity score, consistent with CPIC recommendations.5 A score of 1 was assigned for each normal activity allele (i.e., *1, *2, *35), 0.5 for each decreased activity allele (i.e., *9, *10, *17, *29, *41), and 0 for each no activity allele (i.e., *3–*8, *11, *15). Phenotype was assigned based on the sum of scores (i.e., gene score): 0, PM; 0.5, IM; 1–2, NM; and >2, UM. The genotyping assay also detected allele duplication but not which allele was duplicated or the number of allele copies. Therefore, ranged phenotypes were possible (e.g., NM to UM). Genotype and phenotype results were reported as discrete variables in the EHR in the laboratory results section to enable electronic clinical decision support to alert providers of the PM, IM, or UM phenotype in the event that a CYP2D6-metabolized opioid was prescribed in the future.
A pharmacist refined phenotype assignment for participants taking a CYP2D6 inhibitor using methods described elsewhere21 and referenced in CPIC guidelines.5 Specifically, interacting medications, as defined by the FDA,17 were accounted for by multiplying the gene score by an inhibitor factor, which was 0 for strong (e.g., bupropion, fluoxetine) and 0.5 for moderate (e.g., duloxetine) inhibitors.17 Figure S1 provides an overview for obtaining and applying CYP2D6 genotype data to patient care.
Recommendations based on CYP2D6 phenotype
For participants in the CYP2D6-guided arm, pharmacists provided recommendations to physicians for opioid prescribing based on the assigned CYP2D6 phenotype (Tables S2–S3). Similar to CPIC guidelines,5 recommendations focused on opioids metabolized by CYP2D6 (codeine, tramadol, oxycodone, and hydrocodone) and were for alternative therapy in PMs and UMs secondary to the risk of ineffectiveness and toxicity, respectively. A key difference between CPIC guidelines and recommendations provided in this trial was that recommendations were also made for alternative therapy in IMs taking codeine, tramadol, oxycodone, and hydrocodone if pain was not well controlled. The recommendation was delivered to the prescriber via a clinical consult note in the EHR within 2 weeks after the result returned or prior to the next patient visit. The prescriber was notified of the consult note via email or an in-basket message within the EHR, per their preference. To maintain a pragmatic approach, ultimate prescribing decisions were left to the provider. This allowed the physician to account for other factors important in care (e.g., control of pain on current therapy). Genotype results were not actively reported to patients as part of the study.
Data collection and outcome measures
Baseline data collected consisted of clinical data and patient-reported outcomes via the Pain Collaborative Health Outcomes Information Registry (CHOIR).22 Medication and Pain-CHOIR data were also collected at 3 months. Patients completed the baseline Pain-CHOIR questionnaire on an electronic device (or the study coordinator read the questionnaire from the device) in the clinic at the time of enrollment or by telephone on the same day or approximately one week after the clinic visit. Similarly, the 3-month data were collected in clinic, or if the patient did not have a clinic visit at the 3-month time point, via telephone. Pain-CHOIR utilized computer adaptive testing (CAT), which delivered select questionnaire items based on previous responses to minimize question burden while maintaining test reliability. Pain-CHOIR collected patient-reported pain intensity, opioid-related aberrant behavior,23 pain catastrophizing,24 and the following Patient Reported Outcomes Measurement Information System (PROMIS®) measures: anger, anxiety, depression, fatigue, pain behavior, pain interference, physical function, sleep disturbance, and sleep-related impairment.25 Medication data were collected from the EHR.
Pain intensity was measured in four domains: current, worst, least, and average. The latter three were reflective of pain over the past week. All pain intensity measurements are reported on a scale from 0 to 10, with 10 indicating the highest level of pain. The composite pain intensity was defined as the mean of the pain intensity current, worst, and average. This is similar to “characteristic pain intensity” seen elsewhere in the literature, but on a 0–10 scale.26
The primary outcome was change in the composite pain intensity from baseline to 3 months in patients with the IM or PM phenotype (IM/PMs) treated at baseline with tramadol or codeine. This is the group with the greatest potential for benefit with a CYP2D6-guided approach. The secondary outcome used the same group of patients and examined the percent of participants with ≥30% reduction in the composite pain intensity, a marker of a clinically significant pain reduction.27 Outcomes were compared between the CYP2D6-guided and usual care arms. The same analyses were conducted separately for patients treated with hydrocodone, patients treated with oxycodone, and all patients who completed the 3-month follow-up. While underpowered in these analyses, we sought to gain insight into whether CYP2D6 influenced pain response in these populations.
Implementation metrics examined included the percent of participants in the usual care arm agreeing to genotyping at 3 months and genotype turnaround time. In addition, as a marker of the intervention’s fidelity, changes in pharmacotherapy consistent with phenotype assignment were assessed for each participant. Changes considered consistent with recommendations made in IM/PMs were (1) addition of a nonopioid analgesic in place of an opioid metabolized predominately via CYP2D6, (2) addition of an opioid not predominantly metabolized by CYP2D6 (e.g., morphine) in place of an opioid with CYP2D6-mediated metabolism, and (3) removal of a CYP2D6 inhibitor in a participant prescribed an opioid with CYP2D6-mediated metabolism and judged as IM or PM based on drug interactions but NM based on genotype. The NM phenotype may be associated with any type of pharmacotherapy option.
Data analysis
Baseline characteristics were compared between CYP2D6-guided and usual care arms using a two-sample t test, chi-square analysis, or Fisher’s exact test as appropriate. Multiple linear regression was used to compare the change in composite pain intensity between arms. Covariates entered into a stepwise regression model were group assignment, age, sex, race, baseline characteristics (medical history, type and duration of pain, opioid and nonopioid analgesic use) that differed between arms with P < 0.2 on univariate analysis and baseline patient-reported outcomes (i.e., composite pain intensity, pain catastrophizing, opioid-related aberrant behavior, and PROMIS® measures) also differing between arms with P < 0.2. The five pain intensity dimensions (composite, current, worst, least, and average) exhibited collinearity; therefore, the baseline pain intensity dimensions that did not match the outcome variable in the model were excluded. Covariates with P < 0.05 remained in the final linear regression model. P values generated from multiple linear regression are denoted as adjusted P values (adj P).
The methodology for the primary analysis was repeated for secondary analyses of other pain intensity dimensions (current, worst, least, and average). All pain intensity dimensions were assessed for different patient groups described above. A P < 0.05 was considered statistically significant. Secondary outcomes were not adjusted for multiple comparisons and findings are considered exploratory.
One of our goals was to assess the feasibility of conducting a study of genotype-guided pain management to inform a larger trial. As such, sample size was one of convenience and based on the expected number of patients who could be recruited over the course of the trial, and the power calculation was performed post hoc rather than a priori. With 45 IM/PMs prescribed tramadol or codeine at baseline (CYP2D6-guided n = 29; usual care n = 16), there was 80% power to detect an effect size of ≥0.89, which is equivalent to a difference in pain intensity score of 1.25 (or 18% difference in score), assuming an average pain intensity of 6.9 and standard deviation of 1.4. This represents adequate power as these differences are smaller than previous descriptions of a clinically meaningful difference (~30%).28,29 All analyses were conducted using SAS version 9.4 (SAS, Cary, NC).
RESULTS
Study participants
A total of 370 of 375 (99%) participants completed baseline measures (Fig. 1), and their characteristics are shown in Table 1. The mean age of study participants was 59 years, and most were of European ancestry (71%), with chronic back or arthritis pain (93%). The mean composite pain intensity of the study population was 6.55 of 10 (i.e., moderate to high level of pain),29 and most were prescribed an opioid at baseline (94%); tramadol was most commonly prescribed (45%). Seventy-three percent were on a single opioid, and 20% had two or three opioids listed on their baseline medication profile. Baseline differences between the CYP2D6-guided and usual care arms included race, duration and type of pain, opioid prescribed, nonopioid analgesic use, and history of emotional disorder.
A total of 345 participants (93%) completed 3-month follow-up (Fig. 1). Their CYP2D6 phenotypes based on genotype were similar in frequency to those previously reported (Table 2), and differed by race (Table S4).5 The percent of participants with a CYP2D6 IM or PM phenotype increased from 10% to 35% after drug interactions were considered, and was similar between Whites and Blacks (Table S4). The most commonly prescribed inhibitors causing phenoconversion were duloxetine (n = 47), bupropion (n = 30), fluoxetine (n = 14), and paroxetine (n = 7).
Change in pain intensity
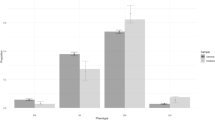
One participant with incomplete PROMIS® measure data was excluded. Analysis proceeded in the remaining 344 participants. Change in pain intensity composite among IM/PMs prescribed tramadol or codeine at baseline favored the CYP2D6-guided group compared with the usual care group (-1.01 ± 1.59 vs. -0.40 ± 1.20; adj P = 0.016; Fig. 2; Table S5; covariates for adjusted analyses are listed in Tables S5–S6). Reductions in pain intensity composite were similar between IMs and PMs (data not shown). Within the same subgroup of IM/PMs prescribed tramadol or codeine at baseline, significantly more participants in the CYP2D6-guided group experienced a 30% reduction in composite pain intensity (7/29 [24%]) compared with the usual care group (0/16 [0%]; P = 0.040). Contrarily, among NMs prescribed tramadol or codeine at baseline, there was no difference in the change in composite pain intensity at 3 months (-0.61 ± 1.39 vs. -0.54 ± 1.69; adj P = 0.540; Fig. 2; Table S5) or the percent with a 30% reduction in composite pain intensity (9/51 [18%] vs. 11/64 [17%]; P = 0.949) between CYP2D6-guided and usual care groups, respectively. In the overall population (all CYP2D6 phenotypes combined) treated with tramadol and codeine at baseline, there was no difference in the composite pain intensity between CYP2D6-guided (-0.72 ± 1.46) and usual care (-0.41 ± 1.71; adj P = 0.078) groups, though there were trends for benefit of a CYP2D6-guided approach.
Represents the change in composite pain intensity from baseline to 3 months for participants with complete follow-up. These data represent 29 CYP2D6-guided and 16 usual care participants with the IM/PM phenotype and 51 CYP2D6-guided and 64 usual care participants with the NM phenotype. Covariates adjusted for in the analysis can be found in Table S5. IM intermediate metabolizer, NM normal metabolizer, PM poor metabolizer.
The hydrocodone-treated IM/PMs exhibited trends in pain improvement between CYP2D6-guided and usual care groups similar to that observed with tramadol and codeine. In a post hoc analysis of 70 IM/PMs prescribed hydrocodone, tramadol, or codeine at baseline, the CYP2D6-guided group had a greater reduction in composite pain intensity (-0.84 ± 1.51) compared with the usual care group (-0.12 ± 1.32; adj P = 0.019; Fig. 3, Table S6). There was no difference in the percent with a 30% reduction in composite pain intensity in the CYP2D6-guided (8/51 [16%]) versus usual care (0/19 [0%]; P = 0.097) groups. Among NMs prescribed hydrocodone, tramadol, or codeine at baseline, the change in composite pain intensity (-0.41 ± 1.51 and -0.34 ± 1.79; adj P = 0.447; Fig. 3, Table S6) and percent with a 30% reduction in composite pain intensity (13/81 [16%] and 11/76 [14%]; P = 0.784) were similar in the CYP2D6-guided and usual care groups, respectively.
Change in composite pain intensity at 3 months for participants prescribed hydrocodone, tramadol, or codeine at baseline. Represents the change in composite pain intensity from baseline to 3 months for participants with complete follow-up. These data represent 51 CYP2D6-guided and 19 usual care participants with the IM/PM phenotype and 81 CYP2D6-guided and 76 usual care participants with the NM phenotype. Covariates adjusted for in the analysis can be found in Table S6. IM intermediate metabolizer, NM normal metabolizer, PM poor metabolizer.
In contrast to the findings for tramadol, codeine, and hydrocodone, among the 47 IM/PMs prescribed oxycodone at baseline, the CYP2D6-guided group experienced less improvement in composite pain intensity compared with the usual care group (-0.02 ± 1.09 vs. -0.87 ± 0.67; adj P = 0.024), respectively. There was no difference in the percent with a 30% reduction in the composite pain intensity in CYP2D6-guided (4/44 [7%]) and usual care (1/10 [9%]) groups (P > 0.99). There was no difference in change in composite pain intensity in oxycodone-treated NMs in the CYP2D6-guided (-0.05 ± 1.56) and usual care (-0.25 ± 1.31) arms (adj P = 0.140).
Data for pain intensities worst, least, average and current are reported in Supplemental Tables S5–S6.
Implementation metrics
Of the 128 participants in the control group, 126 (98%) agreed to clinical genotyping on study completion. The median genotype turnaround time was 8 (interquartile range [IQR] 7–13) days. Among participants in the CYP2D6-guided arm, changes in pharmacotherapy consistent with IM and PM phenotypes occurred more often in IM/PMs (23%, 21/93) compared with NMs (9%, 10/116; P = 0.005). The most common change was addition of nonopioid therapy in place of an opioid metabolized by CYP2D6 (n = 16/21 [76%]). Within the IM/PM subgroup in the guided arm taking tramadol or codeine at baseline, there was no significant change in composite pain intensity between patients with a medication change (per EHR review) and those without (p = 0.692); though it is possible that changes to over-the-counter (OTC) therapy were made, but we were unable to capture that information from the EHR. In the usual care arm, changes in pharmacotherapy were similar across phenotype groups (10% [3/29] of IM/PM vs. 8% [7/92] of NM participants; P = 0.641).
DISCUSSION
This trial demonstrated successful implementation of CYP2D6-guided opioid therapy in the primary care setting and improved pain control in the population hypothesized to benefit most from this approach (i.e., IM/PMs prescribed tramadol or codeine). To our knowledge, this is the first prospective trial to examine pain control with CYP2D6-guided opioid prescribing.
The implementation was deemed successful as changes in pharmacotherapy consistent with recommendations made for IM/PM phenotypes were exhibited in IM/PMs but not NMs in the CYP2D6-guided group. In other words, actions consistent with CYP2D6-guided recommendations occurred more often where expected (IM/PM versus NM). The usual care group did not show differences in prescribing between IM/PM and NM participants, which implies the changes in pharmacotherapy in the CYP2D6-guided participants were due to the intervention. Other indicators of success include the efficient genotype turnaround and high acceptance of genotyping on study completion in the usual care arm.
In the CYP2D6-guided group, IM/PMs prescribed tramadol/codeine at baseline were more likely to have a clinically significant (i.e., ≥30%) reduction in pain intensity27 compared with the usual care arm. Together with the moderate to high pain level participants reported at baseline (6.55/10), our results suggest that future trials of CYP2D6-guided therapy should focus on patients with poorly controlled pain taking an opioid metabolized by CYP2D6. Moreover, the benefit of CYP2D6-guided therapy in IM/PMs was large enough to result in a change in composite pain intensity (P = 0.078) that approached significance in all participants prescribed tramadol or codeine at baseline. Pharmacogenetic trials will require large sample sizes to detect overall population benefits, given that only a portion of the population can benefit from genotype-guided therapy.30,31 The trends for benefit in the overall population (despite not being powered for such) are encouraging regarding the potential population benefit for such an approach in an appropriately powered study.
Some have indicated concern that CPIC guidelines may lead to overuse of strong opioids (e.g., morphine).32 Our clinicians were more likely to move to a nonopioid when presented with a recommendation to avoid CYP2D6-metabolized opioids in IM/PMs. To our knowledge, this is the first assessment of clinician utilization of CYP2D6-guided therapy in the adult chronic pain population, and it suggests potential for this approach to reduce opioid use. Ineffective pain control is the most commonly reported motivation for patients to misuse opioids.33 A precision medicine approach to chronic pain management, with switching to nonopioid therapy in CYP2D6 IMs/PMs, may be a promising strategy to manage pain while helping to combat the opioid crisis.
Oxycodone-treated participants in our trial had less improvement in pain intensity in the CYP2D6-guided versus usual care arm. It is possible that greater pain severity in oxycodone-treated patients contributed to this finding. Nonetheless, our findings are consistent with previous studies in oxycodone-treated patients that have shown no effect of CYP2D6 genotype or use of CYP2D6 inhibitors on pain control, despite lower oxymorphone-to-oxycodone concentrations in PMs versus NMs,34,35,36 suggesting that oxymorphone formation may not be critical for analgesic effects. In contrast, among hydrocodone-treated IM/PMs, we observed trends toward reduced pain intensity in the CYP2D6-guided versus usual care arm, suggesting that, similar to codeine and tramadol, CYP2D6 phenotype is important for hydrocodone response. This is consistent with previous evidence of reduced hydrocodone effectiveness in individuals taking CYP2D6 inhibitors.37
The pragmatic design led to important strengths in regard to external validity. First, because of the limited exclusion criteria, our results are generalizable to adults with varying etiologies of chronic pain managed in primary care clinics. The positive findings are encouraging because most patients (91%) experienced pain for over a year, and some would consider pain of this duration to be difficult to improve. Second, consistent with other reports,38 use of CYP2D6 inhibitors causing phenoconversion was high, which is likely a reflection of the high prevalence of comorbidities often treated with medications that inhibit CYP2D6 (e.g., depression, neuropathy) among patients with chronic pain. Third, like clinical practice, all prescribing decisions were at the physician’s discretion. This allowed for assessing the feasibility of CYP2D6 genotyping in clinical practice and its potential to advise therapy changes. Lastly, participants were receptive to genotyping as seen by near-universal genotyping acceptance in the usual care group and low loss to follow-up in both groups.
Limitations of this trial include the nonrandomized, cluster design, which resulted in baseline differences between arms. However, significant differences in pain improvement were observed after accounting for baseline differences. A future study randomized at the patient level would be important to help ensure groups are well matched. Secondly, given the pragmatic design, patients were not blinded to group assignment. Had this biased patient-reported outcomes, we would have anticipated improvements in pain intensity across phenotype groups in the CYP2D6-guided arm. Improvements in pain intensity in IM/PMs but not NMs increases our confidence that results were not secondary to confounding factors (e.g., placebo effect, difference in clinics). To minimize a placebo effect, results were not actively provided to patients. However, we cannot exclude that the prescriber discussed genotype results with the patient or that the patient viewed results through the patient portal of the EHR, which may have impacted pain scores. This is a natural limitation of a pragmatic design. The benefits of a pragmatic design however are that the external validity for outcomes in a real world setting is high. A third limitation is the difficulty in assigning phenotype for heterozygous genotypes with copy-number variation (CNV) given the inability to determine which allele was duplicated. For example, for the *1/*4 genotype with CNV, the phenotype could range from NM (if *4 was duplicated) to UM (if *1 was multiplicated). Another limitation is that assessment of medications was through EHR review, and did not include assessment of OTC analgesics (e.g., nonsteroidal anti-inflammatory drugs [NSAIDs]). Given that in many cases when therapy was changed, it was to a nonopioid, the prescriber may have recommended that the patient use an OTC NSAID (perhaps at prescription level doses). We would not have captured this information and would have missed that a drug change was made. We also did not collect information on opioid formulation (i.e., short or long acting) and cannot comment on whether results varied by formulation or use of combination therapy with a short- and long-acting opioid. Lastly, the median 8-day genotype turnaround time likely contributed to lower rates of adherence to pharmacist recommendations given that physicians may not act on results until patients return to clinic, which in some cases was after the 3-month study period. It is possible that this trial underestimates the potential benefit of CYP2D6-guided prescribing in IMs/PMs, particularly if a preemptive approach to genotyping is utilized.
In summary, the implementation of CYP2D6-guided care was shown to be feasible and yielded clinically relevant improvements in pain control among the subset of patients most expected to benefit. These results indicate CYP2D6 genotype testing may be a helpful addition to the primary care physician’s armamentarium in chronic pain management. A larger trial to further document the potential benefits of this precision medicine approach is warranted.
References
Nahin RL. Estimates of pain prevalence and severity in adults: United States, 2012. J Pain. 2015;16:769–780.
Levy B, Paulozzi L, Mack KA, Jones CM. Trends in opioid analgesic-prescribing rates by specialty, U.S., 2007-2012. Am J Prev Med. 2015;49:409–413.
Madras BK. The surge of opioid use, addiction, and overdoses: responsibility and response of the US health care system. JAMA Psychiatry. 2017;74:441–442.
IMS Institute for Healthcare Informatics. Medication use and the shifting cost of healthcare—a review of use of medicines in the United States in 2013. http://www.imshealth.com/deployedfiles/imshealth/Global/Content/Corporate/IMS%20Health%20Institute/Reports/Secure/IIHI_US_Use_of_Meds_for_2013.pdf. Accessed August 2014.
Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95:376–382.
Volpe DA, McMahon Tobin GA, Mellon RD, et al. Uniform assessment and ranking of opioid mu receptor binding constants for selected opioid drugs. Regul Toxicol Pharmacol. 2011;59:385–390.
Eckhardt K, Li S, Ammon S, et al. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain. 1998;76:27–33.
Lotsch J, Rohrbacher M, Schmidt H, et al. Can extremely low or high morphine formation from codeine be predicted prior to therapy initiation? Pain. 2009;144:119–124.
Poulsen L, Arendt-Nielsen L, Brosen K, Sindrup SH. The hypoalgesic effect of tramadol in relation to CYP2D6. Clin Pharmacol Ther. 1996;60:636–644.
Stamer UM, Musshoff F, Kobilay M, et al. Concentrations of tramadol and O-desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther. 2007;82:41–47.
Lotsch J, Skarke C, Schmidt H, et al. Evidence for morphine-independent central nervous opioid effects after administration of codeine: contribution of other codeine metabolites. Clin Pharmacol Ther. 2006;79:35–48.
Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. Codeine, ultrarapid-metabolism genotype, and postoperative death. N Engl J Med. 2009;361:827–828.
Gasche Y, Daali Y, Fathi M, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351:2827–2831.
Stamer UM, Stuber F, Muders T, Musshoff F. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth Analg. 2008;107:926–929.
Orliaguet G, Hamza J, Couloigner V, et al. A case of respiratory depression in a child with ultrarapid CYP2D6 metabolism after tramadol. Pediatrics. 2015;135:e753–755.
Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19:69–76.
US Food and Drug Administration. Drug development and drug interactions: table of substrates, inhibitors and inducers. https://www.fda.gov/drugs/developmentapprovalprocess/developmentresources/druginteractionslabeling/ucm093664.htm. Accessed 16 November 2018.
Radford H, Fitzgerald P, Martin S, Johnson MI. A service improvement project to review prescribing information provided by general practitioners for new referrals to a UK National Health Service hospital pain clinic: potential implications of CYP2D6 enzyme inhibition. Br J Pain. 2016;10:222–231.
Tirkkonen T, Laine K. Drug interactions with the potential to prevent prodrug activation as a common source of irrational prescribing in hospital inpatients. Clin Pharmacol Ther. 2004;76:639–647.
Loudon K, Treweek S, Sullivan F, et al. The PRECIS-2 tool: designing trials that are fit for purpose. BMJ. 2015;350:h2147.
Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol. 2010;50:450–458.
Sturgeon JA, Dixon EA, Darnall BD, Mackey SC. Contributions of physical function and satisfaction with social roles to emotional distress in chronic pain: a Collaborative Health Outcomes Information Registry (CHOIR) study. Pain. 2015;156:2627–2633.
Webster LR, Webster RM. Predicting aberrant behaviors in opioid-treated patients: preliminary validation of the Opioid Risk Tool. Pain Med. 2005;6:432–442.
Sullivan MJL, Bishop SR, Pivik J. The Pain Catastrophizing Scale: development and validation. Psychol Assess. 1995;7:524–532.
Cella D, Riley W, Stone A, et al. The Patient-Reported Outcomes Measurement Information System (PROMIS) developed and tested its first wave of adult self-reported health outcome item banks: 2005-2008. J Clin Epidemiol. 2010;63:1179–1194.
Von Korff M, Ormel J, Keefe FJ, Dworkin SF. Grading the severity of chronic pain. Pain. 1992;50:133–149.
Dworkin RH, Turk DC, Wyrwich KW, et al. Interpreting the clinical importance of treatment outcomes in chronic pain clinical trials: IMMPACT recommendations. J Pain. 2008;9:105–121.
Farrar JT, Berlin JA, Strom BL. Clinically important changes in acute pain outcome measures: a validation study. J Pain Symptom Manage. 2003;25:406–411.
Farrar JT, Pritchett YL, Robinson M, Prakash A, Chappell A. The clinical importance of changes in the 0 to 10 numeric rating scale for worst, least, and average pain intensity: analyses of data from clinical trials of duloxetine in pain disorders. J Pain. 2010;11:109–118.
Coenen MJ, de Jong DJ, van Marrewijk CJ, et al. Identification of patients with variants in TPMT and dose reduction reduces hematologic events during thiopurine treatment of inflammatory bowel disease. Gastroenterology. 2015;149:907–917 e907.
Newman WG, Payne K, Tricker K, et al. A pragmatic randomized controlled trial of thiopurine methyltransferase genotyping prior to azathioprine treatment: the TARGET study. Pharmacogenomics. 2011;12:815–826.
Nicholson WT, Formea CM. Clinical perspective on the Clinical Pharmacogenetics Implementation Consortium Updated 2014 guidelines for CYP2D6 and codeine. Clin Chem. 2015;61:319–321.
Han B, Compton WM, Blanco C, et al. Prescription opioid use, misuse, and use disorders in U.S. adults: 2015 National Survey on Drug Use and Health. Ann Intern Med. 2017;167:293–301.
Zwisler ST, Enggaard TP, Mikkelsen S, Brosen K, Sindrup SH. Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand. 2010;54:232–240.
Andreassen TN, Eftedal I, Klepstad P, et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur J Clin Pharmacol. 2012;68:55–64.
Lemberg KK, Heiskanen TE, Neuvonen M, et al. Does co-administration of paroxetine change oxycodone analgesia: an interaction study in chronic pain patients. Scand J Pain. 2017;1:24–33.
Monte AA, Heard KJ, Campbell J, et al. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad Emerg Med. 2014;21:879–885.
Knisely MR, Carpenter JS, Draucker CB, et al. CYP2D6 drug-gene and drug-drug-gene interactions among patients prescribed pharmacogenetically actionable opioids. Appl Nurs Res. 2017;38:107–110.
Acknowledgements
Funded by National Institute of Health (NIH) grants U01 HG007269, UL1TR000064, and institutional support from the University of Florida.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
L.C. has received research funding from Mallinckrodt Pharmaceuticals, the US Food and Drug Administration, and Canon Biomedical. She receives compensation as a reviewer from Wolters Kluwer and the American College of Clinical Pharmacy. C.A.H.'s research has been funded, through grants to his employer organizations, by the National Institutes of Health, Agency for Healthcare Research and Quality, Patient-Centered Outcomes Research Institute, and Pfizer, Inc. C.A.H. has received compensation for grant review with the Agency for Healthcare Research and Quality and medical educational activities for Impact Education, LLC. J.A.J. and R.B.F. have received research funding from the National Institutes of Health. K.N. is an employee of University of Florida Health Pathology Laboratories. P.S. receives support from University of Florida Health Pathology Laboratories. The other authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
At the time of the trial and data collection, author was a postgraduate trainee with the UF Health Personalized Medicine Program
Supplementary information
Rights and permissions
About this article
Cite this article
Smith, D., Weitzel, K.W., Elsey, A.R. et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet Med 21, 1842–1850 (2019). https://doi.org/10.1038/s41436-018-0431-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0431-8
Keywords
This article is cited by
-
Whole blood hydroxychloroquine: Does genetic polymorphism of cytochrome P450 enzymes have a role?
Clinical and Experimental Medicine (2023)
-
Determining the potential clinical value of panel-based pharmacogenetic testing in patients with chronic pain or gastroesophageal reflux disease
The Pharmacogenomics Journal (2021)
-
A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management
Genetics in Medicine (2021)
-
Appraisal and development of evidence-based clinical decision support to enable perioperative pharmacogenomic application
The Pharmacogenomics Journal (2021)
-
CYP2D6 genotype and reduced codeine analgesic effect in real-world clinical practice
The Pharmacogenomics Journal (2021)