Abstract
Purpose
Contiguous gene deletions are known to cause several neurodevelopmental syndromes, many of which are caused by recurrent events on chromosome 16. However, chromosomal microarray studies (CMA) still yield copy-number variants (CNVs) of unknown clinical significance. We sought to characterize eight individuals with overlapping 205-kb to 504-kb 16p13.3 microdeletions that are distinct from previously published deletion syndromes.
Methods
Clinical information on the patients and bioinformatic scores for the deleted genes were analyzed.
Results
All individuals in our cohort displayed developmental delay, intellectual disability, and various forms of seizures. Six individuals were microcephalic and two had strabismus. The deletion was absent in all 13 parents who were available for testing. The area of overlap encompasses seven genes including TBC1D24, ATP6V0C, and PDPK1 (also known as PDK1). Bi-allelic TBC1D24 pathogenic variants are known to cause nonsyndromic deafness, epileptic disorders, or DOORS syndrome (deafness, onychodystrophy, osteodystrophy, mental retardation, seizures). Sanger sequencing of the nondeleted TBC1D24 allele did not yield any additional pathogenic variants.
Conclusions
We propose that 16p13.3 microdeletions resulting in simultaneous haploinsufficiencies of TBC1D24, ATP6V0C, and PDPK1 cause a novel rare contiguous gene deletion syndrome of microcephaly, developmental delay, intellectual disability, and epilepsy.
Similar content being viewed by others
Introduction
Chromosomal microarray (CMA) technology has facilitated the discovery of multiple new microdeletion syndromes previously invisible on conventional karyotypes. However, classification of small deletions as pathogenic can be challenging. Many genes are still poorly characterized and functional data are often unavailable. Therefore, collecting a group of individuals with phenotypic and cytogenetic data can aid in the interpretation of a copy-number variant (CNV), especially for very rare variants.
Autosomal recessive pathogenic variants in TBC1D24 (MIM 613577) lead to epilepsy (familial infantile myoclonic epilepsy (FIME, MIM 605021) early-infantile epileptic encephalopathy 16 (EIEE16, MIM 615338), nonsyndromic hearing loss (either recessive, DFNB86, MIM 614617; or dominant, DFNA65, MIM 616044), or DOORS syndrome (deafness, onychodystrophy, osteodystrophy, mental retardation, and seizures, MIM 220500). We noted that carriers of TBC1D24 pathogenic variants may have a susceptibility to epilepsy notably in the mother of a patient with DOORS syndrome who carries a loss-of-function pathogenic variant,1 and this was eventually noted in other families (detailed in Banuelos et al.2). We thus sought to identify the phenotype associated with microdeletions of TBC1D24 and surrounding genes. We here report on eight individuals with epilepsy and developmental delay who share overlapping microdeletions at 16p13.3 including TBC1D24, ATP6V0C, and PDPK1.
Materials and methods
Cytogenetic laboratories were contacted to identify individuals with microdeletions encompassing TBC1D24. Patients were identified in the cytogenetics laboratories of the institutions where P.M.C. was a faculty member (Baylor College of Medicine) and currently is (Centre hospitalier universitaire [CHU] Sainte-Justine), but also in other centers across the world. Ten individuals had eligible microdeletions and treating clinicians were then approached to recruit patients, and provide clinical details and DNA samples. Eight individuals were enrolled in the study after informed consent was obtained (on consent forms approved by the Baylor College of Medicine and the CHU Sainte-Justine Internal Review Boards). Consent to publish photographs was obtained from each family where a photo is shown. TBC1D24 Sanger sequencing was performed in all individuals except individual 6 (no DNA available) according to published protocols.1 Heterozygous TBC1D24 deletion was confirmed in individual 2 by real-time polymerase chain reaction (PCR) on genomic DNA (data not shown).
Clinical information was collected with a standardized questionnaire. Given the clinical manifestations of DOORS syndrome, specific questions were included on dental anomalies, hearing deficits, dysmorphic facial features, and abnormalities of the hands, nails, and feet. Physicians were asked to provide details on seizure disorders and brain imaging.
CNVs and deleted genes were visualized using the University of California–Santa Cruz (UCSC) genome browser human assembly hg19 (ref. 3). Haploinsufficiency scores (%HI) for the deleted genes were obtained from the DECIPHER database4 (Supplementary Material). Probability of loss-of-function intolerance (pLI) scores were drawn from the ExAC database5 (Supplementary Material). Modeling the probability of autosomal dominant inheritance P(AD) was done with the DOMINO tool6 (Supplementary Material). PubMed, Google Scholar, and OMIM were used for the literature review until February 2018.
Clinical data on individuals
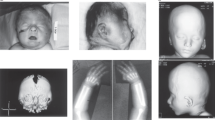
Individual 1 was referred at 8 years for seizures, microcephaly, and developmental delay. She is the only child from a nonconsanguineous union. She was born at term after an uneventful pregnancy. She attends a mainstream school with one-to-one support. Her major difficulties are comprehension and mathematics. At 23 months, she presented with a cluster of generalized tonic–clonic seizures that were treated with levetiracetam and sodium valproate. She has been seizure-free on levetiracetam monotherapy for 5 years. At 5 years, a magnetic resonance image (MRI) was reported as normal. At 8 years, her height and weight were at the 9th percentile, while her head circumference (HC) measured 1.5 cm below the 0.4th percentile for age. She was not dysmorphic (Fig. 1a, b).
Individual 2 came to the attention of a neurometabolic clinic at the age of 6 years. He was born at term to nonconsanguineous parents. Early on, he was noted to have feeding difficulties, failure to thrive, and microcephaly with increased tone. At 13 months, he presented with seizures including generalized tonic–clonic and atonic seizures and head drops. His early developmental milestones were met normally, but at 6 years, he was not yet toilet trained, his speech was limited to single words, and he was able to follow simple verbal commands. He attended kindergarten in an inclusion classroom and received speech, physical, occupational, and applied behavior analysis therapy. His physical exam was remarkable for short stature (<5th percentile) and a HC at the 2nd to 5th percentile. There was no dysmorphism.
Individual 3 is the first of three brothers of nonconsanguineous parents. He was born at term after a normal pregnancy. At birth, length, weight, and HC were at the 25th percentile. During early childhood, he was found to have hyperacusis, hypotonia, and developmental delays (sitting at 9 months, walking after 21 months of age). He attends a specialized classroom. At 30 months, he developed myoclonic astatic epilepsy and was subsequently hospitalized for epileptic encephalopathy. He was treated with valproic acid and lamotrigine. He has been seizure-free since the age of 5 years with normalization of electroencephalogram (EEG) patterns resulting in the discontinuation of valproic acid. On physical exam at 8.5 years, he had microcephaly, prognathism, small teeth with only two permanent teeth, and tapering fingers.
Individual 4 is a 15-year-old male who was born at term to nonconsanguineous parents. He had speech delay and significant learning difficulties. At 15 years, IQ testing (score 51–62) confirmed mild intellectual disability. He exhibits sexualized behavior and has a diagnosis of autism spectrum disorder (ASD) and attention deficit hyperactivity disorder (ADHD). From 15 months, he had convulsions consisting of generalized tonic–clonic seizures that were initially associated with febrile illnesses. From 2 years, he was treated with valproic acid; later clobazam and sulthiame were added. At 5.3 years, his height and weight were above the 90th percentile. He was microcephalic with a HC at the 2nd percentile, but otherwise without dysmorphic features. A neurological exam was normal, including an EEG and an MRI of the brain.
Individual 5 is a 21-year-old male with intellectual disability. At 13 years, he scored below the 1st percentile on the Wechsler Intelligence Scale for Children (WISC-IV). At 10 months, he was diagnosed with generalized tonic–clonic seizures, later he also had episodes of absence and myoclonic or atonic seizures. He has been seizure-free for more than 1 year on a combination treatment of levetiracetam, rufinamide, and clonazapam. An MRI at 13 years revealed a small tubular structure in the right frontal lobe that was interpreted as a normal venous variant. His HC measured at the 2nd percentile at 14 years, with height and weight at the 3rd percentile at 17 years. Mildly dysmorphic features included posteriorly rotated ears and a pointed chin (Fig. 1c, d).
Individual 6 is a 39-year-old man with intellectual disability and significant emotional behavioral concerns with mania and bipolar episodes necessitating multiple psychiatric hospitalizations. From age 3 years, he had generalized tonic–clonic seizures that have been well controlled with the exception of breakthrough seizures at 14 and 25 years. He is treated with phenytoin, buspirone, lorazepam, clonazepam, lamotrigine, olanzapine, and zonisamide. He had corrective surgery for strabismus and multiple dental operations. A brain MRI at 31 years was significant for microcephaly with a thickening of the calvarium and minimal vermian atrophy, which may be secondary to chronic phenytoin use. On physical exam, he had normal height and weight, a tubular nose, and slightly enlarged testicles (Fig. 1e, f).
Individual 7 was born after a normal pregnancy to healthy nonconsanguineous parents. He was diagnosed with hearing loss, strabismus (Fig. 1g, h), and nystagmus with normal vision at 2 years. At 5.5 years, his developmental status was estimated at about 2 years; formal testing was unsuccessful. He is treated for ADHD. Since the age of 13 months, he suffered from generalized tonic–clonic seizures that are moderately controlled with oxcarbazepine, levetiracetam, and valproic acid. MRIs at 2.5 and 4.5 years demonstrated stable cerebral and cerebellar atrophy. At 5.5 years, he was of normal height and weight with a HC at the 2nd to 5th percentile.
Individual 8 was born at 32 weeks estimated gestational age via Caesarean section for nonreassuring fetal heart tracing. At birth, her height and weight measured at the 10th percentile, whereas head growth was preserved at the 50th percentile. At 6.5 years, she measured at the 10th percentile for height and weight with a HC below the 3rd percentile. Gross motor and language development is delayed and her IQ was measured at 58 with the Culture Fair Intelligence Test (CFT-R). She experienced her first febrile seizure at 18 months, followed by a cluster of febrile and afebrile tonic seizures at 20 months and 2.4 years. She experienced two more seizure clusters of myoclonic seizures lasting up to 7 days requiring polytherapy of valproic acid, clobazam, and levetiracetam and has been seizure-free on this combination for 2 years. On physical exam, she has a high forehead, a long tubular nose with a broad nasal ridge, and epicanthal folds.
For more information, see Supplementary Material.
Results
Clinical and cytogenetic data were available on eight individuals (Table 1, Supplementary Material). All eight suffered from childhood-onset epilepsy, mostly generalized tonic–clonic seizures (six individuals). All eight individuals also have variable developmental delays ranging from mild to moderate and affecting speech, and fine and gross motor skills, with three being diagnosed with ADHD and one with ASD. Cranial MRI findings were normal for five individuals and nonspecific in three. Interestingly, some features observed in this cohort, such as microcephaly (six individuals), hypotonia (two individuals), hearing loss (one individual), and visual impairment (two individuals), have been previously associated with biallelic TBC1D24 pathogenic variants. Four individuals had mild dysmorphic features (Table 1, Fig. 1). The three Caucasian individuals for whom images are available (Fig. 1) share facial similarities such as a sloping forehead, a long tubular nose with a prominent columella, and a prominent chin.
CMA identified overlapping microdeletions on the short arm of chromosome 16 (16p13.3; Fig. 2). There is no overlap with the 16p13.3 (refs. 7,8) and 16p11.2 (refs. 9,10) deletion syndromes. The smallest deletion (individual 8) contains 13 genes and the largest (individual 3) contains 25 genes (Supplementary Table 1). Parental testing in six families determined the deletion to be a de novo event. For individual 1, her tested mother is not a carrier. In individual 3, the deletion was present in 83% of cells, suggesting a postzygotic event. The deletions do not share a common break point and range in size from 205 kb to 504 kb with a minimally overlapping region (MOR) of 112 kb that includes seven genes (UCSC genome browser hg19) TBC1D24 (MIM 613577), ATP6V0C (MIM 108745), AMDHD2 (amidohydrolase domain-containing 2), CEMP1 (Cementum protein 1, MIM 611113), MIR3168 (microRNA 3168), PDPK1 (or PDK1, 3-phosphoinositide dependent protein kinase-1, MIM 605213), and DQ577714 (piRNA38825).
We next looked at bioinformatic prediction scores. A %HI score of less than 10% is predictive of haploinsufficiency of a heterozygously deleted gene. A pLI score of ≥0.9 is indicative of intolerance to loss-of-function pathogenic variants and haploinsufficiency. A P(AD) of ≥0.95 is highly associated with autosomal dominant inheritance through haploinsufficiency, gain-of-function or dominant-negative effects. Of the genes within the MOR, PDPK1 reaches the lowest %HI at 27% and the highest pLI score at 0.95. DOMINO predicts PDPK1 to “very likely” cause autosomal dominant conditions with a P(AD) of 0.986. However, none of the genes in the MOR reach significant %HI scores of less than 10% (Table 2). Complete Sanger sequencing of the nondeleted TBC1D24 allele did not detect any pathogenic variants and therefore excludes an AR epilepsy phenotype in this cohort (data not shown).
Discussion
Several factors favor a causative link between microdeletions at 16p13.3 and the clinical manifestations in this group. The phenotype is very homogeneous with all individuals suffering from epilepsy and variable degrees of developmental delay. In addition, the majority is microcephalic and none have additional malformations or major medical problems. In all six for whom this data were available, the deletion occurred de novo. Furthermore, CNVs containing the MOR have not been identified in normal controls in several large-scale studies.11,12,13 Only one additional case with a comparable deletion was found in a cohort of 29,085 cases with intellectual disability, developmental delay, and/or ASD, but clinical information is not available (see supplemental table 7 in ref. 12). The microdeletion was absent in two additional cohorts, 1 of 5531 cases that were sent to a diagnostic laboratory for clinical testing14 and 1 including 1133 children with severe developmental disorders.15
Our results suggest that 16p13.3 microdeletions encompassing TBC1D24, ATP6V0C, and PRPK1 genes represent a novel contiguous gene deletion epileptic syndrome. TBC1D24, a known epilepsy gene, encodes a member of the Tre2-Bub2-Cdc16 (TBC) domain-containing RAB-specific GTPase-activating proteins. Analysis of the crystal structure of the Drosophila ortholog Skywalker (Sky) identified a cationic pocket that is preserved in human TBC1D24. This pocket is necessary for binding to the lipid membrane via phosphoinositides phosphorylated at the 4 and 5 positions. Abrogation of the cationic pocket by introduction of two human TBC1D24 pathogenic variants found in DOORS syndrome led to impaired synaptic vesicle trafficking and seizures in Drosophila.16 TBC1D24 is the only gene in the MOR that is associated with autosomal dominant and recessive human disease phenotypes.
ATP6V0C (ATPase, H+ transporting, lysosomal 16 kDa, V0 subunit C) is a component of vacuolar ATPase (V-ATPase), a multisubunit enzyme that mediates acidification of eukaryotic intracellular organelles. It is present in endosomes, lysosomes, clathrin-coated vesicles, and the Golgi complex, where it is essential to acidification and maintenance of endocytic and exocytic pathways.17 Experiments in zebrafish embryos suggest a neuron-specific expression of the zebrafish ortholog atp6v0c2 where it is associated with presynaptic vesicles and involved in neurotransmitter storage.18
PDPK1 (also known as PDK1) is a highly conserved protein kinase that is involved in many different signaling pathways (reviewed in ref. 19). Similar to TBC1D24, it is able to bind to phosphatidylinositol 3,4,5-trisphosphate or phosphatidylinositol 3,4-bisphosphate produced at the plasma membrane where it fulfills an important function in cell migration.20 While homozygous Pdpk1 knockout mice die on embryonic day E9.5 (ref. 21), mice with residual PDK1 activity (10–30%) are viable and fertile, albeit of a smaller size.22 The reduced interaction of PDPK1 with phosphoisonitides leads to a decrease in PKB/mTORC1/BRSK signaling, decreased neuronal cell size in vivo, and shorter cortical neuron length in vitro.23 To date, evidence on direct interactions between the three main genes of interest has not been published.
Other genes in the MOR are less likely to play a causative role in the pathogenesis of this recurrent deletion. The enzyme AMDHD2 is involved in a degradation pathway that tightly regulates N-glycolylneuraminic acid (Neu5Gc),24 a protein that is incorporated at low levels into the surface glycoproteins of several human tissues.25 However, loss-of-function pathogenic variants of metabolic disorders are usually well tolerated in the carrier state. Cementum protein 1 (CEMP1) is a marker of cementoblast-related cells and plays a role in cementoblast differentiation in periodontal ligament. It is not expressed in brain.26 Expression studies in hepatocellular carcinoma (HCC) suggest a role of MIR-3178 as a tumor suppressor by inhibiting cell proliferation, angiogenesis, invasion, and migration of HCC tumor endothelial cells.27 The potential role of MIR-3178 in other organ systems and during development has not yet been studied. For DQ577714, to date, no investigations detailing the function of its gene product have been published.
While individuals with recessive TBC1D24 pathogenic variants have more severe phenotypes than our cohort, in some families with recessive epilepsy or DOORS syndrome, carriers or obligate carriers also suffered from a milder form of childhood epilepsy.1,2,28,29 In the ExAC database, the number of expected loss-of-function (LoF) variants (n = 10.7) corresponds to the number of observed LoF variants (n = 10) for TBC1D24, which seems to contradict our suggestion that haploinsufficiency for TBC1D24 may predispose to epilepsy. However, it is important to note that the incidence of epilepsy is relatively high in the general population (7 per 1000 [ref. 30]) and the ExAC data set only excludes severe childhood-onset disorders. It is therefore possible that some TBC1D24 heterozygous LoF or deleterious missense variants may lower the threshold for the development of mild forms of epilepsy in some families. In animal studies, Tbc1d24 has been shown to be important for neuronal migration and cortical maturation by facilitating the transition of migrating neurons into a bipolar shape.31 PDPK1 is also involved in neuronal differentiation in mice.23 The third candidate gene within the MOR, ATP6V0C, like TBC1D24, can regulate vesicular trafficking. While heterozygous Atp6v0c knockout mice are phenotypically normal,32 homozygous embryos develop only to the blastocyst stage and die shortly after implantation.33
In recent years, several exome sequencing studies have been conducted in patient cohorts with severe epilepsy, developmental delays, or both who often remained undiagnosed after a standard genetic evaluation with CMA and targeted gene sequencing.15,34-39 Different de novo frameshift variants in ATP6V0C were found in one individual in a study performing exome sequencing of 80 patients with Dravet syndrome34 and in 1 individual from a cohort of 4293 families undergoing exome sequencing for severe developmental delay.39 Details on their phenotypes were not provided and the variants were not validated by functional assays. In the Dravet syndrome study, the authors conducted targeted sequencing of ATP6V0C in 67 additional families and did not identify other pathogenic variants. One proband in a cohort of 1133 children with severe developmental delay was found to have a de novo missense variant in PDPK1 by exome sequencing, but no phenotype information was provided (Table S2 in ref. 15). De novo variants in either gene were absent from other studies with cohort sizes ranging from 50 to 293 trios35,37,38 and none of the above-cited studies listed de novo variants in TBC1D24. Neither of the three genes emerged as a strong individual candidate gene for either severe epilepsy or developmental delay in these studies; however, further large-scale cohort studies or functional assays are needed to explore the possible contribution of PDPK1 and ATP6V0C LoF variants to developmental delay and epilepsy phenotypes.
In conclusion, while haploinsufficiency of TBC1D24, ATP6V0C, or PDPK1 may be tolerated individually (larger cohorts will be useful to provide a definitive answer), our results suggest that haploinsufficiency for a combination of these genes leads to developmental delay and epilepsy as observed in this cohort. Future studies are needed to further refine the MOR and elucidate the individual and cumulative effect of the genes implicated in this phenotype.
Change history
14 January 2019
The original version of this Article contained an error in the spelling of the author Siddharth Banka, which was incorrectly given as Siddhart Banka. This has now been corrected in both the PDF and HTML versions of the Article.
References
Campeau PM, Kasperaviciute D, Lu JT, et al. The genetic basis of DOORS syndrome: an exome-sequencing study. Lancet Neurol. 2014;13:44–58.
Banuelos E, Ramsey K, Belnap N. et al. Case report: novel mutations in TBC1D24 are associated with autosomal dominant tonic-clonic and myoclonic epilepsy and recessive Parkinsonism, psychosis, and intellectual disability. F1000Res. 2017;6:553.
Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006.
Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285.
Quinodoz M, Royer-Bertrand B, Cisarova K, Di Gioia SA, Superti-Furga A, Rivolta C. DOMINO: using machine learning to predict genes associated with dominant disorders. Am J Hum Genet. 2017;101:623–629.
Bartsch O, Rasi S, Delicado A, et al. Evidence for a new contiguous gene syndrome, the chromosome 16p13.3 deletion syndrome alias severe Rubinstein-Taybi syndrome. Hum Genet. 2006;120:179–186.
Nelson M, Quinonez S, Ackley T, Iyer RK, Innis JW. Multiple congenital anomalies and developmental delay in a boy associated with a de novo 16p13.3 deletion. Am J Med Genet A. 2011;155A:612–617.
Ghebranious N, Giampietro PF, Wesbrook FP, Rezkalla SH. A novel microdeletion at 16p11. 2 harbors candidate genes for aortic valve development, seizure disorder, and mild mental retardation. Am J Med Genet A. 2007;143:1462–1471.
Bijlsma E, Gijsbers A, Schuurs-Hoeijmakers J, et al. Extending the phenotype of recurrent rearrangements of 16p11. 2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87.
Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838.
Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063.
Männik K, Mägi R, Macé A, et al. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313:2044–2054.
Vulto‐van Silfhout AT, Hehir‐Kwa JY, Bon BW, et al. Clinical significance of de novo and inherited copy‐number variation. Hum Mutat. 2013;34:1679–1687.
Fitzgerald T, Gerety S, Jones W, et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223.
Fischer B, Lüthy K, Paesmans J, et al. Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function. Nat Struct Mol Biol. 2016;23:965.
Mangieri LR, Mader BJ, Thomas CE, et al. ATP6V0C knockdown in neuroblastoma cells alters autophagy-lysosome pathway function and metabolism of proteins that accumulate in neurodegenerative disease. PLoS ONE. 2014;9:e93257.
Chung AY, Kim MJ, Kim D, et al. Neuron‐specific expression of atp6v0c2 in zebrafish CNS. Dev Dyn. 2010;239:2501–2508.
Gagliardi PA, di Blasio L, Primo L. PDK1: A signaling hub for cell migration and tumor invasion. Biochim Biophys Acta. 2015;1856:178–188.
di Blasio L, Gagliardi PA, Puliafito A, et al. PDK1 regulates focal adhesion disassembly by modulating endocytosis of αvβ3 integrin. J Cell Sci. 2015;128:863–877.
Lawlor MA, Mora A, Ashby PR, et al. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002;21:3728–3738.
Bayascas JR, Wullschleger S, Sakamoto K, et al. Mutation of the PDK1 PH domain inhibits protein kinase B/Akt, leading to small size and insulin resistance. Mol Cell Biol. 2008;28:3258–3272.
Zurashvili T, Cordón-Barris L, Ruiz-Babot G, et al. Interaction of PDK1 with phosphoinositides is essential for neuronal differentiation but dispensable for neuronal survival. Mol Cell Biol. 2013;33:1027–1040.
Bergfeld AK, Pearce OM, Diaz SL, Pham T, Varki A. Metabolism of vertebrate amino sugars with N-glycolyl groups: elucidating the intracellular fate of the non-human sialic acid N-glycolylneuraminic acid. J Biol Chem. 2012;287:28865–28881.
Diaz SL, Padler-Karavani V, Ghaderi D, et al. Sensitive and specific detection of the non-human sialic acid N-glycolylneuraminic acid in human tissues and biotherapeutic products. PLoS ONE. 2009;4:e4241.
Alvarez-Pérez MA, Narayanan S, Zeichner-David M, Carmona BR, Arzate H. Molecular cloning, expression and immunolocalization of a novel human cementum-derived protein (CP-23). Bone. 2006;38:409–419.
Li W, Shen S, Wu S, Chen Z, Hu C, Yan R. Regulation of tumorigenesis and metastasis of hepatocellular carcinoma tumor endothelial cells by microRNA-3178 and underlying mechanism. Biochem Biophys Res Commun. 2015;464:881–887.
Zara F, Gennaro E, Stabile M, et al. Mapping of a locus for a familial autosomal recessive idiopathic myoclonic epilepsy of infancy to chromosome 16p13. Am J Hum Genet. 2000;66:1552–1557.
Balestrini S, Milh M, Castiglioni C, et al. TBC1D24 genotype–phenotype correlation: epilepsies and other neurologic features. Neurology. 2016;87:77–85.
Hirtz D, Thurman D, Gwinn-Hardy K, Mohamed M, Chaudhuri A, Zalutsky R. How common are the “common” neurologic disorders? Neurology. 2007;68:326–337.
Falace A, Buhler E, Fadda M, et al. TBC1D24 regulates neuronal migration and maturation through modulation of the ARF6-dependent pathway. Proc Natl Acad Sci U S A. 2014;111:2337–2342.
Inoue H, Noumi T, Nagata M, Murakami H, Kanazawa H. Targeted disruption of the gene encoding the proteolipid subunit of mouse vacuolar H+-ATPase leads to early embryonic lethality. Biochim Biophys Acta. 1999;1413:130–138.
Sun-Wada G-H, Murata Y, Yamamoto A, Kanazawa H, Wada Y, Futai M. Acidic endomembrane organelles are required for mouse postimplantation development. Dev Biol. 2000;228:315–325.
Carvill GL, Weckhuysen S, McMahon JM, et al. GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology. 2014;82:1245–1253.
Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344.
Helbig KL, Hagman KDF, Shinde DN, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016;18:898.
Bowling KM, Thompson ML, Amaral MD, et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017;9:43.
Hamdan FF, Myers CT, Cossette P, et al. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet. 2017;101:664–685.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Mucha, B.E., Banka, S., Ajeawung, N.F. et al. A new microdeletion syndrome involving TBC1D24, ATP6V0C, and PDPK1 causes epilepsy, microcephaly, and developmental delay. Genet Med 21, 1058–1064 (2019). https://doi.org/10.1038/s41436-018-0290-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0290-3
Keywords
This article is cited by
-
ATP6V0C gene variants were identified in individuals with epilepsy, with or without developmental delay
Journal of Human Genetics (2023)
-
Genetic architecture and phenotypic landscape of deafness and onychodystrophy syndromes
Human Genetics (2022)
-
The Ncoa7 locus regulates V-ATPase formation and function, neurodevelopment and behaviour
Cellular and Molecular Life Sciences (2021)