Abstract
Recent dramatic advances in multiomics research coupled with exponentially increasing volume, complexity, and interdisciplinary nature of publications are making it challenging for scientists to stay up-to-date on the literature. Strategies to address this challenge include the creation of online databases and warehouses to support timely and targeted dissemination of research findings. Although most of the early examples have been in cancer genomics and pharmacogenomics, the approaches used can be adapted to support investigators in heart, lung, blood, and sleep (HLBS) disorders research. In this article, we describe the creation of an HLBS population genomics (HLBS-PopOmics) knowledge base as an online, continuously updated, searchable database to support the dissemination and implementation of studies and resources that are relevant to clinical and public health practice. In addition to targeted searches based on the HLBS disease categories, cross-cutting themes reflecting the ethical, legal, and social implications of genomics research; systematic evidence reviews; and clinical practice guidelines supporting screening, detection, evaluation, and treatment are also emphasized in HLBS-PopOmics. Future updates of the knowledge base will include additional emphasis on transcriptomics, proteomics, metabolomics, and other omics research; explore opportunities for leveraging data sets designed to support scientific discovery; and incorporate advanced machine learning bioinformatics capabilities.
Similar content being viewed by others
Introduction
“Upon this gifted age, in its dark hour,
Rains from the sky a meteoric shower
Of facts… they lie unquestioned, uncombined.
Wisdom enough to leech us of our ill is daily spun;
but there exists no loom to weave it into fabric.”
― Edna St. Vincent Millay (1939).1
Dramatic advances in basic and early translational research in genomics, epigenomics, transcriptomics, proteomics, metabolomics, pharmacogenomics, and genetic epidemiology coupled with the exponentially increasing volume and interdisciplinary nature of publications are making it difficult for individual scientists to keep up with the literature in a timely fashion.2,3,4 To address this challenge, several strategies5,6,7,8,9 have been used to create online databases and warehouses10,11,12,13,14 for timely dissemination of research findings and for managing the data deluge.9 Most of the early examples have been in cancer genomics, pharmacogenomics, and public health genomics.15,16,17,18 For example, the Office of Public Health Genomics at the Centers for Disease Control and Prevention (CDC) developed the Public Health Genomics Knowledge Base (PHGKB) as an online, continuously updated, searchable database of published scientific literature to disseminate information on and track the impact of genomics on population health.18
Within the National Heart, Lung, and Blood Institute (NHLBI) mission areas, no such resource exists as a one-stop online site to systematically identify, curate, and expediently disseminate published genomics research findings that are essential for continued population science research and practice for reducing the burden of heart, lung, blood, and sleep (HLBS) diseases and disorders. The recent NHLBI Strategic Vision emphasis on genomics and other “omics” research19 and important advances in the NHLBI Trans-Omics and Precision Medicine (TOPMed) program20 creates an opportunity for rapid advances in our understanding of HLBS pathobiological processes and how they impact individual and population health. Most importantly, a freely accessible online knowledge base in HLBS diseases is needed to help accelerate the dissemination and implementation of genomics research findings that are relevant to clinical and public health practice, as well as leverage related NHLBI resources and the expertise of TOPMed program investigators. To address this need, we have created HLBS-PopOmics by building on the foundations of the PHGKB.18
Materials and methods
Details of the architecture, contents, methods, and early results of PHGKB have been published previously.6,18,21 In essence, its core content includes Web-based curated scientific resources, especially PubMed references and abstracts for epidemiologic, translational and implementation studies captured by weekly horizon scanning and indexed and grouped into thematic categories. Machine learning techniques and text mining were used to facilitate automatic data screening and collection while online back-end expert-screening and data entry pipelines ensured readiness and efficiency of the manual data collection and data entry processes. The system retrieves PubMed abstracts of scientific publications from PubMed with Medical Subject Headings (MeSH) indexing combined with the incorporation of Unified Medical Language System (UMLS) controlled vocabularies.22 PHGKB was built on open source architecture and platform for the software development using J2EE technology and other Java open source frameworks, including Hibernate and Strut as described by Yu et al.18
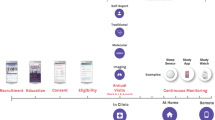
We categorize search findings into the four thematic areas of heart, lung, blood, or sleep disorders as shown in Fig. 1. Selected examples of the diseases and conditions under these four thematic areas that constitute the NHLBI mission areas are provided in Fig. 1 and a link to an alphabetical list of diseases, conditions, tests, and procedures in the NHLBI health topics (Appendix).23 In addition, two cross-cutting areas are identified to include (1) the Ethical, Legal, and Social Implications (ELSI) of Genomics Research; and (2) Evidence Reviews, Clinical Practice Guidelines, and advances in Implementation Science that have relevance for population genomics.
To populate HLBS-PopOmics with relevant data from two key PHGKB databases (Genomics & Health Impact Weekly Scan Database and Human Genome Epidemiology Literature Finder Database), a back-end script automatically searches and extracts data based on the common diseases, conditions, tests, and procedures in the NHLBI health topics23 (Appendix). Data from the Weekly Scan Database is further categorized into six subcategories to yield information on cross-cutting themes shown in Table 1 (Translation Research/Implementation Studies, Evidence Synthesis, Guidelines, Reviews/Commentaries, Tools/Methods, and Ethical/Legal and Social Issues).
Results
The landing page of HLBS-PopOmics displays the most common diseases/disorders for each HLBS category, with user-friendly quick links listed on the Common Health Topics panel. As of 13 March 2018, there are 175 health topics related to HLBS that are assigned to the four main categories (Heart and Vascular Diseases, Lung Diseases, Blood Disorders, and Sleep Disorders). Free text search capability is also provided for users to explore any HLBS topic(s) of interest. Pertinent information for each disease category or specific search term is presented in the “What’s New” section, which displays the top 15 records from different databases based on the data entry time. The Information in Specialized Databases section gathers statistics from different databases, categories, and reference information, and presents the data in a tabulated format for easy navigation. Under each tab, the top five records for a given database or categorized information is displayed, and the “More” link will lead to a complete data set in the individual database interface.
To date, the majority of the information in the HLBS-PopOmics database (~87%) comes from epidemiologic studies (Table 2). Of the HLBS diseases/disorders focal areas, heart and vascular diseases comprise the most content in all six of the data types within the Genomics & Health Impact Scan Database. Sleep disorders, on the other hand, contain the least information of all the disease areas, highlighting a research area that has significant growth potential (Table 2). There have been steady yearly increases in the number of Translation and Implementation Studies and Reviews/Commentaries since 2012. However, there has not been a significant change in the number of publications centered on evidence synthesis, tools/methods, or guidelines since 2014 (Fig. 2). While epidemiological studies remain the largest data source in HLBS-PopOmics, the number of publications has significantly decreased since 2015. Genome-wide association studies (GWAS) do not follow the same trend and remain minimally changed since 2012 (Fig. 3).
Discussion
Effective strategies to support HLBS investigators, policy makers, and practitioners to stay up-to-date on the relevance of multiomics research for clinical practice through timely and targeted dissemination of research advances are needed. These strategies are also important in efforts to accelerate the dissemination and implementation of genomics research advances to reduce the population burden of HLBS disorders. Knowledge bases such as PHGKB that have been designed to address this challenge can also help bridge population-based research on genomics with clinical and public health applications.18 HLBS-PopOmics leverages the current framework and infrastructure of PHGKB and lessons learned from PHGKB specialty applications in cancer24 and infectious diseases25 to provide timely and targeted dissemination of multiomics research advances to support HLBS research investigators with special emphasis on advancing implementation science.26,27
As noted in Fig. 1, ethical, legal, and social implications (ELSI) issues are key cross-cutting areas within HLBS genomics, and across the entire genomics field. These issues also align with an ELSI-specific genomic research area.28 Challenging ELSI issues include understanding the best way to communicate just-in-time information that may predict disease and health, uncertainty about the implications of genetic variants, and what interventions may be beneficial. A broad range of ELSI research areas involve examining the various impacts of science and technology on society. Strategies to foster ELSI-related research and practical insights on how a scientific research team should incorporate a strong and effective ELSI program within the broader genomics research mandate are much needed.28 Topics currently include ELSI and biobanking, changing legal landscapes, screening, informed choice and decision making, privacy and anonymity, and issues related to youth in the United States and globally.
Strengths
A major strength of HLBS-PopOmics is its ability to display quickly, for any health topic relevant to heart, lung, blood, and sleep disorders, the state of genomic science translation to clinical practice and population health impact. Thus, HLBS-PopOmics can be used by researchers for rapidly identifying gaps in translation and implementation research in a particular subject area, and by practitioners for finding relevant guidelines, tools, and implementation programs to guide clinical and public health practice. In the rapidly moving world of genomics and related fields, the overwhelming research is still in scientific discoveries and early translation (bench to bedside). HLBS-PopOmics makes it easier to find quickly the 1–2% of the relevant scientific literature, guidelines, and evidence syntheses that are relevant to current clinical practice. Expert curation and machine learning procedures associated with PHGKB help researchers and practitioners alike to find and track over time, genomic scientific resources in the T2–T4 translation space.29 In addition, users can customize their searches using the MyPHGKB feature of the knowledge base by choosing specific topics of interests and databases to search, as well as receive automatic updates by email.
For example, a quick search (12 March 2018) for familial hypercholesterolemia (FH), a common genetic disorder associated with premature heart disease reveals information and publications relevant to clinical practice today, including classification of cascade screening for FH as a tier 1 application (ready for implementation and T4 research), 263 epidemiologic studies of the prevalence, genes and clinical outcomes of FH in various populations, 22 papers synthesizing the evidence base using meta analyses and economic analyses, 18 clinical guidelines, and 125 translation and implementation studies showing implementation and impact of FH in clinical practice in the real world. The search results also provide valuable embedded linkages to related resources such as the National Institutes of Health (NIH) genetic testing registry, the Pharmacogenomics Knowledgebase (PharmGKB), OMIM, and NHLBI resources such as TOPMed.
Limitations
The data in PHGKB in general is collected by the horizon scanning method developed by the Centers for Disease Control and Prevention (CDC) Office of Public Health Genomics6 for translational research including a PubMed targeted search query, supplemented by monitoring of online news using Google Alerts, and genomics-related websites; and a machine learning technology21 for human genome epidemiological studies. The completeness of the data collection in the areas is always a challenging task for the databases. For example, the PHGKB includes translational research studies from 2012 on, as well as epidemiologic studies since 2001; thus, publications and other information prior to these years are not present.
The data in the aggregation levels (e.g., all HLBS and four areas) are heavily dependent on the health topic list provided in the system, which might not be complete, including all possible synonyms of each topic term. Although much of the scientific literature in the databases has been indexed with the US National Library of Medicine’s Medical Subject Headings (MeSH), which have been used in the information retrieval with adoption of UMLS, there are still a significant number of publications without MeSH indexing. The efficiency of information retrieval might be impacted by this shortcoming.
Future
We anticipate that future revisions of HLBS-PopOmics will include an expansion of HLBS diseases, conditions, tests, and procedures as the list of NHLBI health topics increases. In addition, future versions will introduce search terms that go beyond disease categories and reflect cross-cutting themes such as circadian biology. Future versions will also include additional emphasis on transcriptomics, proteomics, metabolomics, and other omics research as well as explore opportunities for leveraging data sets designed to support scientific discovery. We will continue to do usability testing in the near future, especially as we change or add new features to the knowledge base. Importantly, future revisions of HLBS-PopOmics will benefit from emerging advances in new tools such as supervised machine learning30,31,32 and other automated computational methodologies and bioinformatics capabilities.33,34,35
Conclusions
In summary, HLBS-PopOmics allows researchers, policy makers, and practitioners to stay up-to-date on the rapidly moving developments in genomics and related fields. Most importantly, it allows users to rapidly assess the relevance of current and emerging public health and population genomics research findings in the prevention, detection, evaluation, treatment, and control of heart, lung, blood, and sleep disorders.
References
Millay ESV. Huntsman, What Quarry? Sonnets of Edna St. Vincent Millay. 1st ed. New York: Harper & Brothers; 1939.
Landhuis E. Scientific literature: information overload. Nature. 2016;535:457–458.
Khare R, Leaman R, Lu Z. Accessing biomedical literature in the current information landscape. Methods Mol Biol . 2014;1159:11–31.
Khoury MJ, Gwinn M, Clyne M, Yu W. Genetic epidemiology with a capital E, ten years after. Genet Epidemiol . 2011;35:845–852.
Gwinn M, Grossniklaus DA, Yu W, et al. Horizon scanning for new genomic tests. Genet Med. 2011;13:161–165.
Clyne M, Schully SD, Dotson WD, et al. Horizon scanning for translational genomic research beyond bench to bedside. Genet Med. 2014;16:535–538.
Dotson WD, Douglas MP, Kolor K, et al. Prioritizing genomic applications for action by level of evidence: a horizon-scanning method. Clin Pharmacol Ther. 2014;95:394–402.
Wilhite SE, Barrett T. Strategies to explore functional genomics data sets in NCBI’s GEO database. Methods Mol Biol . 2012;802:41–53.
Gobeill J, Pasche E, Vishnyakova D, Ruch P. Managing the data deluge: data-driven GO category assignment improves while complexity of functional annotation increases. Database. 2013;2013:bat041.
Huang K, Brady A, Mahurkar A, et al. MetaRef: a pan-genomic database for comparative and community microbial genomics. Nucleic Acids Res. 2014;42(database issue):D617–624.
Sulakhe D, Balasubramanian S, Xie B, et al. Lynx: a database and knowledge extraction engine for integrative medicine. Nucleic Acids Res . 2014;42(database issue):D1007–1012.
Saito K, Arai S, Kato H. A nutrigenomics database--integrated repository for publications and associated microarray data in nutrigenomics research. Br J Nutr. 2005;94:493–495.
Lyne M, Smith RN, Lyne R, et al. metabolicMine: an integrated genomics, genetics and proteomics data warehouse for common metabolic disease research. Database. 2013;2013:bat060.
Park YK, Bang OS, Cha MH, et al. SigCS base: an integrated genetic information resource for human cerebral stroke. BMC Syst Biol. 2011;5(suppl 2):S10.
Perez-Llamas C, Gundem G, Lopez-Bigas N. Integrative cancer genomics (IntOGen) in Biomart. Database. 2011;2011:bar039.
Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov . 2012;2:401–404.
Sarris K, Komianou A, Patrinos GP, Katsila T. Application of the DruGeVar database in cancer genomics and pharmacogenomics. Public Health Genomics. 2017;20:142–147.
Yu W, Gwinn M, Dotson WD, et al. A knowledge base for tracking the impact of genomics on population health. Genet Med. 2016;18:1312–1314.
National Heart, Lung, and Blood Institute. Charting the future together: the NHLBI strategic vision. Bethesda, MD: NHLBI; 2016.
National Heart, Lung, and Blood Institute. Trans-Omics for Precision Medicine (TOPMed) Program. 2016. https://www.nhlbi.nih.gov/research/resources/nhlbi-precision-medicine-initiative/topmed. August 20, 2018.
Yu W, Clyne M, Dolan SM, et al. GAPscreener: an automatic tool for screening human genetic association literature in PubMed using the support vector machine technique. BMC Bioinformatics. 2008;9:205 https://doi.org/10.1186/1471-2105-9-205.:205-209
Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32:281–291.
National Heart, Lung, and Blood Institute. Health Topics. 2018. https://www.nhlbi.nih.gov/health-topics. August 20, 2018.
Centers for Disease Control and Prevention. Cancer Genomics: Cancer PHGKB. 2018. https://phgkb.cdc.gov/PHGKB/specificPHGKB.action?topic=cancer&query=home. August 20, 2018.
Centers for Disease Control and Prevention. Infectious Diseases: Infectious Diseases PHGKB. 2018. https://phgkb.cdc.gov/PHGKB/specificPHGKB.action?topic=Infectious%20diseases&query=home. Accessed 17 February 2018.
National Academies of Sciences, Engineering, and Medicine. Applying an implementation science approach to genomic medicine: workshop summary. Washington, DC: The National Academies Press; 2016.
Chambers DA, Feero WG, Khoury MJ. Convergence of implementation science, precision medicine, and the learning health care system: a new model for biomedical research. JAMA. 2016;315:1941–1942.
Pullman D, Etchegary H. Clinical genetic research 3: genetics ELSI (ethical, legal, and social issues) research. Methods Mol Biol. 2015;1281:369–382.
Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, Bradley L. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med. 2007;9:665–674.
Schrider DR, Kern AD. Supervised machine learning for population genetics: a new paradigm. Trends Genet . 2018;34:301–312.
Fabris F, Magalhaes JP, Freitas AA. A review of supervised machine learning applied to ageing research. Biogerontology. 2017;18:171–188.
Holder LB, Haque MM, Skinner MK. Machine learning for epigenetics and future medical applications. Epigenetics. 2017;12:505–514.
Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet . 2015;16:321–332.
Padhukasahasram B. Inferring ancestry from population genomic data and its applications. Front Genet . 2014;5:204.
Yuan X, Miller DJ, Zhang J, Herrington D, Wang Y. An overview of population genetic data simulation. J Comput Biol . 2012;19:42–54.
Acknowledgements
We are grateful to our colleague, Michael Twery, who provided constructive comments on an earlier version of this document.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Disclaimer:The views expressed in this article are those of the authors and do not necessarily represent the views of the National Institutes of Health, the Centers for Disease Control and Prevention, or the United States Department of Health and Human Services.
Rights and permissions
About this article
Cite this article
Mensah, G.A., Yu, W., Barfield, W.L. et al. HLBS-PopOmics: an online knowledge base to accelerate dissemination and implementation of research advances in population genomics to reduce the burden of heart, lung, blood, and sleep disorders. Genet Med 21, 519–524 (2019). https://doi.org/10.1038/s41436-018-0118-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0118-1