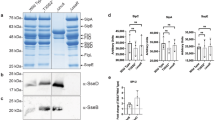
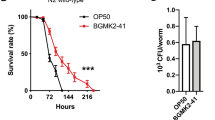
Abstract
Bacterial effector molecules are crucial infectious agents that can cause pathogenesis. In the present study, the pathogenesis of toxic Salmonella enterica serovar Typhi (S. Typhi) proteins on the model host Caenorhabditis elegans was investigated by exploring the host’s regulatory proteins during infection through the quantitative proteomics approach. Extracted host proteins were analyzed using two-dimensional gel electrophoresis (2D-GE) and differentially regulated proteins were identified using MALDI TOF/TOF/MS analysis. Of the 150 regulated proteins identified, 95 were downregulated while 55 were upregulated. The interaction network of regulated proteins was predicted using the STRING tool. Most downregulated proteins were involved in muscle contraction, locomotion, energy hydrolysis, lipid synthesis, serine/threonine kinase activity, oxidoreductase activity, and protein unfolding. Upregulated proteins were involved in oxidative stress pathways. Hence, cellular stress generated by S. Typhi proteins in the model host was determined using lipid peroxidation as well as oxidant and antioxidant assays. In addition, candidate proteins identified via extract analysis were validated by western blotting, and the roles of several crucial molecules were analyzed in vivo using transgenic strains (myo-2 and col-19) and mutant (ogt-1) of C. elegans. To the best of our knowledge, this is the first study to report protein regulation in host C. elegans exposed to toxic S. Typhi proteins. It highlights the significance of p38 MAPK and JNK immune pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
References
Popham JD, Webster JM. Cadmium toxicity in the free-living nematode, Caenorhabditis elegans. Environ Res. 1979;20:183–91.
Hodgkin J, Kuwabara PE, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol. 2000;10:1615–8.
Leung MC, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, et al. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicological Sci. 2008;106:5–28.
Hunt PR. The C. elegans model in toxicity testing. J Appl Toxicol. 2017;1:50–9.
Huffman DL, Bischof LJ, Griffitts JS, Aroian RV. Pore worms: using Caenorhabditis elegans to study how bacterial toxins interact with their target host. Int J Med Microbiol. 2004;293:599–607.
Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol. 2003;13:47–52.
Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–60.
Cezairliyan B, Vinayavekhin N, Grenfell-Lee D, Yuen GJ, Saghatelian A, Ausubel FM. Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 2013;9:e1003101.
Ray A, Rentas C, Caldwell GA, Caldwell KA. Phenazine derivatives cause proteotoxicity and stress in C. elegans. Neurosci Lett. 2015;584:23–7.
Vigneshkumar B, Pandian SK, Balamurugan K. Regulation of Caenorhabditis elegans and Pseudomonas aeruginosa machinery during interactions. Arch Microbiol. 2012;194:229–42.
Kamaladevi A, Balamurugan K. Lipopolysaccharide of Klebsiella pneumoniae attenuates immunity of Caenorhabditis elegans and evades by altering its supramolecular structure. RSC Adv. 2016;6:30070–80.
JebaMercy G, Prithika U, Lavanya N, Sekar C, Balamurugan K. Changes in Caenorhabditis elegans immunity and Staphylococcal virulence factors during their interactions. Gene. 2015;558:159–72.
Kesika P, Karutha Pandian S, Balamurugan K. Analysis of Shigella flexneri-mediated infections in model organism Caenorhabditis elegans. Scand J Infect Dis. 2011;43:286–95.
Durai S, Pandian SK, Balamurugan K. Establishment of a Caenorhabditis elegans infection model for Vibrio alginolyticus. J basic Microbiol. 2011;51:243–52.
JebaMercy G, Vigneshwari L, Balamurugan KA. MAP Kinase pathway in Caenorhabditis elegans is required for defense against infection by opportunistic Proteus species. Microbes Infect. 2013;15:550–68.
Sivamaruthi BS, Balamurugan K. Physiological and immunological regulations in Caenorhabditis elegans infected with Salmonella enterica serovar Typhi. Indian J Microbiol. 2014;54:52–8.
Kamaladevi A, Balamurugan K. Global proteomics revealed Klebsiella pneumoniae induced autophagy and oxidative stress in caenorhabditis elegans by inhibiting PI3K/AKT/mTOR pathway during infection. Front Cell Infect Microbiol. 2017;7:393.
Balasubramanian V, Sellegounder D, Suman K, Krishnaswamy B. Proteome analysis reveals translational inhibition of Caenorhabditis elegans enhances susceptibility to Pseudomonas aeruginosa PAO1 pathogenesis. J Proteom. 2016;145:141–52.
Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol. 2014;4:5. 391
Thaver D, Zaidi AK, Critchley JA, Azmatullah A, Madni SA, Bhutta ZA. Fluoroquinolones for treating typhoid and paratyphoid fever (enteric fever). Cochrane Database of Systematic Reviews. 2008;8:CD004530.
Zaki SA, Karande S. Multidrug-resistant typhoid fever: a review. J Infect Developing Ctries. 2011;5:324–37.
Galan JE. Salmonella interactions with host cells: type III secretion at work. Annu Rev cell developmental Biol. 2001;17:53–86.
Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, et al. Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect Immun. 2015;83:2897–906.
Kao CY, Salwen MJ, Hu SL, Pitter HM, Woollard JM. Diamphidia toxin, the Bushmen’s arrow poison: possible mechanism of prey-killing. Toxicon. 1989;27:1351–66.
Thorne GM, Gorbach SL. General characteristics: nomenclature of microbial toxins. Pharmacol therapeutics. 1981;13:193–203.
Zhang Y, Zou X, Ding Y, Wang H, Wu X, Liang B. Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans. BMC genomics. 2013;14:1–3.
Gavin KA, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–71.
Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sánchez-Pérez P, Cadenas S, et al. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–97.
Schouest K, Zitova A, Spillane C, Papkovsky DB. Toxicological assessment of chemicals using Caenorhabditis elegans and optical oxygen respirometry. Environ Toxicol Chem: Int J. 2009;28:791–9.
Ren H, Zhang H. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Developmental Biol. 2010;345:144–55.
Marsh EK, May RC. Caenorhabditis elegans, a model organism for investigating immunity. Appl Environ Microbiol. 2012;78:2075–81.
Li H, Ren C, Shi J, Hang X, Zhang F, Gao Y, et al. C. A proteomic view of Caenorhabditis elegans caused by short-term hypoxic stress. Proteome Sci. 2010;8:49.
Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–42.
Sem X, Rhen M. Pathogenicity of Salmonella enterica in Caenorhabditis elegans relies on disseminated oxidative stress in the infected host. PLoS One. 2012;7:e45417.
Aksoy D, Şen E. Investigation of pathogenic phenotypes and virulence determinants of food-borne Salmonella enterica strains in Caenorhabditis elegans animal model. Mikrobiyoloji Bul. 2015;49:513–24.
Sahu SN, Anriany Y, Grim CJ, Kim S, Chang Z, Joseph SW, et al. Identification of virulence properties in Salmonella Typhimurium DT104 using Caenorhabditis elegans. PLoS One. 2013;8:e76673.
Wittmann‐Liebold B, Graack HR, Pohl T. Two‐dimensional gel electrophoresis as tool for proteomics studies in combination with protein identification by mass spectrometry. Proteomics. 2006;6:4688–703.
May C, Brosseron F, Chartowski P, Schumbrutzki C, Schoenebeck B, Marcus K. Instruments and methods in proteomics. Data Mining Protÿeomics. 2011;696:3–26.
Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–47.
Prithika U, Deepa V, Balamurugan K. External induction of heat shock stimulates the immune response and longevity of Caenorhabditis elegans towards pathogen exposure. Innate Immun. 2016;22:466–78.
Soti C, Nagy E, Giricz Z, Vígh L, Csermely P, Ferdinandy P. Heat shock proteins as emerging therapeutic targets. Br J Pharmacol. 2005;146:769–80.
Powers MV, Jones K, Barillari C, Westwood I, Montfort RL, Workman P. Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle. 2010;9:1542–50.
Durai S, Singh N, Kundu S, Balamurugan K. Proteomic investigation of Vibrio alginolyticus challenged Caenorhabditis elegans revealed regulation of cellular homeostasis proteins and their role in supporting innate immune system. Proteomics. 2014;14:1820–32.
JebaMercy G, Durai S, Prithika U, Marudhupandiyan S, Dasauni P, Kundu S, et al. Role of DAF-21protein in Caenorhabditis elegans immunity against Proteus mirabilis infection. J Proteom. 2016;145:81–90.
Wang B, Wang H, Xiong J, Zhou Q, Wu H, Xia L, et al. A proteomic analysis provides novel insights into the stress responses of Caenorhabditis elegans towards nematicidal Cry6A Toxin from Bacillus thuringiensis. Sci Rep. 2017;71:1–4.
Mohri-Shiomi A, Garsin DA. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J Biol Chem. 2008;283:194–201.
Balasubramaniam B, Vinitha T, Deepika S, JebaMercy G, VenkataKrishna LM, Balamurugan K. Analysis of Caenorhabditis elegans phosphoproteome reveals the involvement of a molecular chaperone, HSP-90 protein during Salmonella enterica Serovar Typhi infection. Int J Biol Macromol. 2019;137:620–46.
Green RA, Kao HL, Audhya A, Arur S, Mayers JR, Fridolfsson HN, et al. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–82.
Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J Exp Zool Part A: Comp Exp Biol. 2006;305:720–30.
Hellerer T, Axang C, Brackmann C, Hillertz P, Pilon M, Enejder A. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc Natl Acad Sci. 2007;104:14658–63.
Srinivasan S, Sadegh L, Elle IC, Christensen AG, Faergeman NJ, Ashrafi K. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 2008;7:533–44.
Shi X, Tarazona P, Brock TJ, Browse J, Feussner I, Watts JL. A Caenorhabditis elegans model for ether lipid biosynthesis and function. J lipid Res. 2016;57:265–75.
Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochimica et Biophysica Acta (BBA)-Mol Basis Dis. 2012;1822:1442–52.
Kho MF, Bellier A, Balasubramani V, Hu Y, Hsu W, Nielsen-LeRoux C, et al. The pore-forming protein Cry5B elicits the pathogenicity of Bacillus sp. against Caenorhabditis elegans. PLoS One. 2011;6:e29122.
Kasai KI, Hirabayashi J. Galectins: a family of animal lectins that decipher glycocodes. J Biochem. 1996;119:1–8.
Yang RY, Rabinovich GA, Liu FT. Galectins: structure, function and therapeutic potential. Expert Rev Mol Med. 2008;10:e17.
Boscher C, Dennis JW, Nabi IR. Glycosylation, galectins and cellular signaling. Curr Opin Cell Biol. 2011;23:383–92.
Wahlby C, Conery AL, Bray MA, Kamentsky L, Larkins-Ford J, Sokolnicki KL, et al. High-and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods. 2014;683:492–9.
Fouad AD, Pu SH, Teng S, Mark JR, Fu M, Zhang K, et al. Quantitative assessment of fat levels in Caenorhabditis elegans using dark field microscopy. G3: Genes Genomes Genet. 2017;7:1811–8.
Chen WW, Yi YH, Chien CH, Hsiung KC, Ma TH, Lin YC, et al. Specific polyunsaturated fatty acids modulate lipid delivery and oocyte development in C. elegans revealed by molecular-selective label-free imaging. Sci Rep. 2016;6:1–3.
Rogalski TM, Gilbert MM, Devenport D, Norman KR, Moerman DG. DIM-1, a novel immunoglobulin superfamily protein in Caenorhabditis elegans, is necessary for maintaining bodywall muscle integrity. Genetics. 2003;163:905–15.
Otey CA, Rachlin A, Moza M, Arneman D, Carpen O. The palladin/myotilin/myopalladin family of actin‐associated scaffolds. Int Rev Cytol. 2005;246:31–58.
Etheridge T, Oczypok EA, Lehmann S, Fields BD, Shephard F, Jacobson LA, et al. Calpains mediate integrin attachment complex maintenance of adult muscle in Caenorhabditis elegans. PLoS genet. 2012;8:e1002471.
Lecroisey C, Brouilly N, Qadota H, Mariol MC, Rochette NC, Martin E, et al. ZYX-1, the unique zyxin protein of Caenorhabditis elegans, is involved in dystrophin-dependent muscle degeneration. Mol Biol Cell. 2013;24:1232–49.
Luo S, Schaefer AM, Dour S, Nonet ML. The conserved LIM domain-containing focal adhesion protein ZYX-1 regulates synapse maintenance in Caenorhabditis elegans. Development. 2014;141:3922–33.
Stiernagle T. Maintenance of C. elegans. Online Review of C. elegans Biology, wormbook. 1999;2:51–67.
Wingfield P. Protein precipitation using ammonium sulfate. Curr Protoc Protein Sci. 1998;13:A–3F.
Park JW, Lee SG, Song JY, Joo JS, Chung MJ, Kim SC, et al. Proteomic analysis of Helicobacter pylori cellular proteins fractionated by ammonium sulfate precipitation. Electrophoresis. 2008;13:2891–903.
Park GE, Oh HN, Ahn SY. Improvement of the ammonia analysis by the phenate method in water and wastewater. Bull Korean Chem Soc. 2009;30:2032–8.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Ananthi S, Santhosh RS, Nila MV, Prajna NV, Lalitha P, Dharmalingam K. Comparative proteomics of human male and female tears by two-dimensional electrophoresis. Exp Eye Res. 2011;92:454–63.
Sethupathy S, Prasath KG, Ananthi S, Mahalingam S, Balan SY, Pandian SK. Proteomic analysis reveals modulation of iron homeostasis and oxidative stress response in Pseudomonas aeruginosa PAO1 by curcumin inhibiting quorum sensing regulated virulence factors and biofilm production. J Proteom. 2016;145:112–26.
Bianchi L, Driscoll M. Culture of embryonic C. elegans cells for electrophysiological and pharmacological analyses. WormBook. 2006;10:1551–8507.
Scherz‐Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–60.
Wolff SP Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 19941; 233:182–9.
Paoletti F, Mocali A. Determination of superoxide dismutase activity by purely chemical system based on NAD (P) H oOxidation. Methods Enzymol. 1990;186:209–20.
Aebi H. Catalase in vitro in method of enzymology. Methods Enzymol.1984;105:121–26.
Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, et al. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–78.
Ohkawa H, Ohishi N, Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem. 1979;95:351–8.
Chen F, Cushion MT. Use of an ATP bioluminescent assay to evaluate viability of Pneumocystis carinii from rats. J Clin Microbiol. 1994;32:2791–800.
Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, Tran QT, Lawhon S, et al. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect Immun. 2005;73:146–54.
Liu C, Yu X. ADP-ribosyltransferases and poly ADP-ribosylation. Curr Protein Pept Sci. 2015;16:491–501.
O’Rourke B. Mitochondrial ion channels. Annu Rev Physiol. 2007;69:19–49.
Mu K, Wang D, Kitts DD. Molecular mechanisms that define redox balance function in pathogen-host interactions—is there a role for dietary bioactive polyphenols? Int J Mol Sci. 2019;20:6222.
Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2:a004390.
Jeong DE, Lee D, Hwang SY, Lee Y, Lee JE, Seo M, et al. Mitochondrial chaperone HSP‐60 regulates anti‐bacterial immunity via p38 MAP kinase signaling. EMBO J. 2017;36:1046–65.
Acknowledgements
We thank the Caenorhabditis Genetics Centre, which is funded by the National Institute of Health, National Centre for Research Resources for providing the nematode strains. DAM gratefully acknowledges the University Grants Commission (UGC) of India for the financial assistance in the form of UGC-PF (F. No. 42–222/2013 (SR)). The computational facility provided by the Bioinformatics Infrastructure Facility, Alagappa University funded by the Department of Biotechnology, Ministry of Science and Technology, Government of India (Grant No.BT/BI/25/015/2012 (BIF)) are thankfully acknowledged. We are also grateful for the Instrumentation Facility provided by the Department of Science and Technology (DST), Government of India through DST PURSE [Grant No. SR/S9Z-415 23/2010/42(G)], DST FIST [Grant No.SR-FST/LSI-087/2008], UGC through SAP-DRS1 [Grant No. F. 3–28/2011(SAP-II)] and RUSA 2.0. [F. 24–51/2014-U, Policy (TN Multi-Gen), Dept. of Education, GOI].
Author information
Authors and Affiliations
Contributions
DAM, BB and MB designed and performed the experiments and analysed the data. VK helped in proteomic analysis. KB wrote the manuscript in consultation with DAM and other authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Consent for publication
The content of the manuscript has not been published, or submitted for publication elsewhere. All authors have approved the manuscript for submission
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mir, D.A., Balasubramaniam, B., VenkataKrishna, L.M. et al. Proteomic analysis of Caenorhabditis elegans against Salmonella Typhi toxic proteins. Genes Immun 22, 75–92 (2021). https://doi.org/10.1038/s41435-021-00132-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-021-00132-w