Abstract
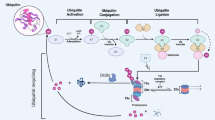
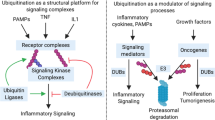
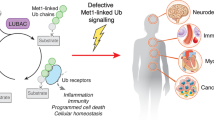
The covalent post-translational modification of proteins by ubiquitination not only influences protein stability and half-life, but also several aspects of protein function including enzymatic activity, sub-cellular localization, and interactions with binding partners. Protein ubiquitination status is determined by the action of large families of ubiquitin ligases and deubiquitinases, whose combined activities regulate many physiological and cellular pathways. The Ubiquitin Specific Protease (USP) family is one of 8 subfamilies of deubiquitinating enzymes composed of more than 50 members. Recent studies have shown that USP15 plays a critical role in regulating many aspects of immune and inflammatory function of leukocytes in response to a broad range of infectious and autoimmune insults and following tissue damage. USP15 regulated pathways reviewed herein include TLR signaling, RIG-I signaling, NF-kB, and IRF3/IRF7-dependent transcription for production of pro-inflammatory cytokines and type I interferons. In addition, USP15 has been found to regulate pathways implicated in tumor onset and progression such as p53, and TGF-β signaling, but also influences the leukocytes-determined immune and inflammatory microenvironment of tumors to affect progression and outcome. Hereby reviewed are recent studies of USP15 in model cell lines in vitro, and in mutant mice in vivo with reference to available human clinical datasets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kaiser P, Huang L. Global approaches to understanding ubiquitination. Genome Biol. 2005;6:233.
Wilkinson KD, Audhya TK. Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J Biol Chem. 1981;256:9235–41.
Brinkmann K, Schell M, Hoppe T, Kashkar H. Regulation of the DNA damage response by ubiquitin conjugation. Front Genet. 2015;6:98.
Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29.
Shembade N, Harhaj EW. Elucidating dynamic protein-protein interactions and ubiquitination in NF-kappaB signaling pathways. Methods Mol Biol. 2015;1280:283–95.
Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86.
Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–14.
Ciechanover A, Elias S, Heller H, Hershko A. “Covalent affinity” purification of ubiquitin-activating enzyme. J Biol Chem. 1982;257:2537–42.
Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–57.
Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21.
Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–7.
Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6.
Ohtake F, Tsuchiya H, Saeki Y, Tanaka K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc Natl Acad Sci USA. 2018;115:E1401–E1408.
Rieser E, Cordier SM, Walczak H. Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem Sci. 2013;38:94–102.
Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45.
Passmore LA, Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem J. 2004;379:513–25.
Wang G, Gao Y, Li L, Jin G, Cai Z, Chao JI, et al. K63-linked ubiquitination in kinase activation and cancer. Front Oncol. 2012;2:5.
Erpapazoglou Z, Walker O, Haguenauer-Tsapis R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells. 2014;3:1027–88.
Lee BL, Singh A, Mark Glover JN, Hendzel MJ, Spyracopoulos L. Molecular basis for K63-linked ubiquitination processes in double-strand DNA break repair: a focus on kinetics and dynamics. J Mol Biol. 2017;429:3409–29.
Carpenter S, O’Neill LA. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem J. 2009;422:1–10.
Lopez-Castejon G, Edelmann MJ. Deubiquitinases: novel therapeutic targets in immune surveillance? Mediators Inflamm. 2016;2016:3481371.
Friend SF, Deason-Towne F, Peterson LK, Berger AJ, Dragone LL. Regulation of T cell receptor complex-mediated signaling by ubiquitin and ubiquitin-like modifications. Am J Clin Exp Immunol. 2014;3:107–23.
Pinto-Fernandez A, Davis S, Schofield AB, Scott HC, Zhang P, Salah E, et al. Comprehensive landscape of active deubiquitinating enzymes profiled by advanced chemoproteomics. Front Chem. 2019;7:592.
Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63.
Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86.
Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 2016;63:146–55.
Kolattukudy PE, Niu J. Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circ Res. 2012;110:174–89.
Hewings DS, Heideker J, Ma TP, AhYoung AP, El Oualid F, Amore A, et al. Reactive-site-centric chemoproteomics identifies a distinct class of deubiquitinase enzymes. Nat Commun. 2018;9:1162.
Haahr P, Borgermann N, Guo X, Typas D, Achuthankutty D, Hoffmann S, et al. ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability. Mol Cell. 2018;70:165–74 e6.
Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2.
Tran HJ, Allen MD, Lowe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry. 2003;42:11460–5.
Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207.
Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–46.
Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–96.
Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7.
Grou CP, Pinto MP, Mendes AV, Domingues P, Azevedo JE. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci Rep. 2015;5:12836.
Pfoh R, Lacdao IK, Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr Relat Cancer. 2015;22:T35–54.
Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–54.
Clerici M, Luna-Vargas MP, Faesen AC, Sixma TK. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat Commun. 2014;5:5399.
Renatus M, Parrado SG, D’Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–302.
Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–56.
Tencer AH, Liang Q, Zhuang Z. Divergence in ubiquitin interaction and catalysis among the ubiquitin-specific protease family deubiquitinating enzymes. Biochemistry. 2016;55:4708–19.
Bonacci T, Emanuele MJ. Dissenting degradation: deubiquitinases in cell cycle and cancer. Semin Cancer Biol. 2020;67:145–58.
Das S, Ramakrishna S, Kim KS. Critical roles of deubiquitinating enzymes in the nervous system and neurodegenerative disorders. Mol Cells. 2020;43:203–14.
Csizmadia T, Low P. The role of deubiquitinating enzymes in the various forms of autophagy. Int J Mol Sci. 2020;21:4196.
Clemente V, D’Arcy P, Bazzaro M. Deubiquitinating enzymes in coronaviruses and possible therapeutic opportunities for COVID-19. Int J Mol Sci. 2020;21:3492.
Kim SY, Baek KH. TGF-beta signaling pathway mediated by deubiquitinating enzymes. Cell Mol Life Sci. 2019;76:653–65.
He M, Zhou Z, Shah AA, Zou H, Tao J, Chen Q, et al. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 2016;6:62.
Chou CK, Chang YT, Korinek M, Chen YT, Yang YT, Leu S, et al. The regulations of deubiquitinase USP15 and its pathophysiological mechanisms in diseases. Int J Mol Sci. 2017;18:483.
Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol. 2014;15:562–70.
Huang X, Langelotz C, Hetfeld-Pechoc BK, Schwenk W, Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J Mol Biol. 2009;391:691–702.
Hayes SD, Liu H, MacDonald E, Sanderson CM, Coulson JM, Clague MJ, et al. Direct and indirect control of mitogen-activated protein kinase pathway-associated components, BRAP/IMP E3 ubiquitin ligase and CRAF/RAF1 kinase, by the deubiquitylating enzyme USP15. J Biol Chem. 2012;287:43007–18.
Das T, Park JK, Park J, Kim E, Rape M, Kim EE, et al. USP15 regulates dynamic protein-protein interactions of the spliceosome through deubiquitination of PRP31. Nucleic Acids Res. 2017;45:4866–80.
Fielding AB, Concannon M, Darling S, Rusilowicz-Jones EV, Sacco JJ, Prior IA, et al. The deubiquitylase USP15 regulates topoisomerase II alpha to maintain genome integrity. Oncogene. 2018;37:2326–42.
Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403.
Lu Y, Qiu Y, Chen P, Chang H, Guo L, Zhang F, et al. ER-localized Hrd1 ubiquitinates and inactivates Usp15 to promote TLR4-induced inflammation during bacterial infection. Nat Microbiol. 2019;4:2331–46.
Zhou Q, Cheng C, Wei Y, Yang J, Zhou W, Song Q, et al. USP15 potentiates NF-kappaB activation by differentially stabilizing TAB2 and TAB3. FEBS J. 2020;287:3165–83.
Schweitzer K, Bozko PM, Dubiel W, Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J. 2007;26:1532–41.
Zou Q, Jin J, Xiao Y, Zhou X, Hu H, Cheng X, et al. T cell intrinsic USP15 deficiency promotes excessive IFN-gamma production and an immunosuppressive tumor microenvironment in MCA-induced fibrosarcoma. Cell Rep. 2015;13:2470–9.
Torre S, Polyak MJ, Langlais D, Fodil N, Kennedy JM, Radovanovic I, et al. USP15 regulates type I interferon response and is required for pathogenesis of neuroinflammation. Nat Immunol. 2017;18:54–63.
Zhang H, Wang D, Zhong H, Luo R, Shang M, Liu D, et al. Ubiquitin-specific protease 15 negatively regulates virus-induced type I interferon signaling via catalytically-dependent and -independent mechanisms. Sci Rep. 2015;5:11220.
Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, et al. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal. 2014;7:ra3.
Chiang C, Pauli EK, Biryukov J, Feister KF, Meng M, White EA, et al. The human papillomavirus E6 oncoprotein targets USP15 and TRIM25 to suppress RIG-I-mediated innate immune signaling. J Virol. 2018;92:e01737–17.
Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–75.
Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med. 2012;18:429–35.
de Jong RN, Ab E, Diercks T, Truffault V, Daniels M, Kaptein R, et al. Solution structure of the human ubiquitin-specific protease 15 DUSP domain. J Biol Chem. 2006;281:5026–31.
Harper S, Besong TM, Emsley J, Scott DJ, Dreveny I. Structure of the USP15 N-terminal domains: a beta-hairpin mediates close association between the DUSP and UBL domains. Biochemistry. 2011;50:7995–8004.
Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–86.
Faesen AC, Luna-Vargas MP, Sixma TK. The role of UBL domains in ubiquitin-specific proteases. Biochem Soc Trans. 2012;40:539–45.
Luna-Vargas MP, Faesen AC, van Dijk WJ, Rape M, Fish A, Sixma TK. Ubiquitin-specific protease 4 is inhibited by its ubiquitin-like domain. EMBO Rep. 2011;12:365–72.
Ward SJ, Gratton HE, Indrayudha P, Michavila C, Mukhopadhyay R, Maurer SK, et al. The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel. J Biol Chem. 2018;293:17362–74.
Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, et al. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol. 2005;15:1217–21.
Zhang Q, Harding R, Hou F, Dong A, Walker JR, Bteich J, et al. Structural basis of the recruitment of ubiquitin-specific protease USP15 by spliceosome recycling factor SART3. J Biol Chem. 2016;291:17283–92.
Zhao B, Velasco K, Sompallae R, Pfirrmann T, Masucci MG, Lindsten K. The ubiquitin specific protease-4 (USP4) interacts with the S9/Rpn6 subunit of the proteasome. Biochem Biophys Res Commun. 2012;427:490–6.
Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, et al. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24:1434–47.
Vlasschaert C, Xia X, Coulombe J, Gray DA. Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol Biol. 2015;15:230.
Baker RT, Wang XW, Woollatt E, White JA, Sutherland GR. Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin-specific protease related to the UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics. 1999;59:264–74.
Angelats C, Wang XW, Jermiin LS, Copeland NG, Jenkins NA, Baker RT. Isolation and characterization of the mouse ubiquitin-specific protease Usp15. Mamm Genome. 2003;14:31–46.
Park KC, Choi EJ, Min SW, Chung SS, Kim H, Suzuki T, et al. Tissue-specificity, functional characterization and subcellular localization of a rat ubiquitin-specific processing protease, UBP109, whose mRNA expression is developmentally regulated. Biochem J. 2000;349:443–53.
Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20.
Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front Immunol. 2018;9:2379.
Marongiu L, Gornati L, Artuso I, Zanoni I, Granucci F. Below the surface: the inner lives of TLR4 and TLR9. J Leukoc Biol. 2019;106:147–60.
Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–51.
Choudhury NR, Heikel G, Michlewski G. TRIM25 and its emerging RNA-binding roles in antiviral defense. Wiley Interdiscip Rev RNA. 2020;11:e1588.
Huang L, Liu H, Zhang K, Meng Q, Hu L, Zhang Y, et al. Ubiquitin-conjugating enzyme 2S enhances viral replication by inhibiting type I IFN production through recruiting USP15 to deubiquitinate TBK1. Cell Rep. 2020;32:108044.
Wang L, Zhao W, Zhang M, Wang P, Zhao K, Zhao X, et al. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J Virol. 2013;87:4507–15.
Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86.
Yaginuma Y, Yoshimoto M, Tokuda A. USP15 inhibits HPV16 E6 degradation and catalytically inactive USP15 has reduced inhibitory activity. Acta Virol. 2018;62:147–56.
Colak S, Ten Dijke P. Targeting TGF-beta signaling in cancer. Trends Cancer. 2017;3:56–71.
Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50:924–40.
Niederkorn M, Hueneman K, Choi K, Varney ME, Romano L, Pujato MA, et al. TIFAB regulates USP15-mediated p53 signaling during stressed and malignant hematopoiesis. Cell Rep. 2020;30:2776–90 e6.
Iyengar PV, Jaynes P, Rodon L, Lama D, Law KP, Lim YP, et al. USP15 regulates SMURF2 kinetics through C-lobe mediated deubiquitination. Sci Rep. 2015;5:14733.
Liu WT, Huang KY, Lu MC, Huang HL, Chen CY, Cheng YL, et al. TGF-beta upregulates the translation of USP15 via the PI3K/AKT pathway to promote p53 stability. Oncogene. 2017;36:2715–23.
Jiang B, Zhou L, Lu J, Wang Y, Liu C, Liang Z, et al. Clinicopathological and prognostic significance of ubiquitin-specific peptidase 15 and its relationship with transforming growth factor-beta receptors in patients with pancreatic ductal adenocarcinoma. J Gastroenterol Hepatol. 2020;36:507–15.
Al-Salihi MA, Herhaus L, Macartney T, Sapkota GP. USP11 augments TGFbeta signalling by deubiquitylating ALK5. Open Biol. 2012;2:120063.
Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-beta type I receptor. Nat Cell Biol. 2012;14:717–26.
Oikonomaki M, Bady P, Hegi ME. Ubiquitin specific peptidase 15 (USP15) suppresses glioblastoma cell growth via stabilization of HECTD1 E3 ligase attenuating WNT pathway activity. Oncotarget. 2017;8:110490–502.
Chen LL, Smith MD, Lv L, Nakagawa T, Li Z, Sun SC, et al. USP15 suppresses tumor immunity via deubiquitylation and inactivation of TET2. Sci Adv. 2020;6:eabc9730.
van den Berk P, Lancini C, Company C, Serresi M, Sanchez-Bailon MP, Hulsman D, et al. USP15 deubiquitinase safeguards hematopoiesis and genome integrity in hematopoietic stem cells and leukemia cells. Cell Rep. 2020;33:108533.
Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014;23:5227–42.
Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–6.
Ge P, Dawson VL. Dawson TMPINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson’s disease. Mol Neurodegener. 2020;15:20.
Cornelissen T, Vilain S, Vints K, Gounko N, Verstreken P, Vandenberghe W. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. Elife. 2018;7:e35878.
Minter MR, Moore Z, Zhang M, Brody KM, Jones NC, Shultz SR, et al. Deletion of the type-1 interferon receptor in APPSWE/PS1DeltaE9 mice preserves cognitive function and alters glial phenotype. Acta Neuropathol Commun. 2016;4:72.
Roy ER, Wang B, Wan YW, Chiu G, Cole A, Yin Z, et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J Clin Invest. 2020;130:1912–30.
Main BS, Zhang M, Brody KM, Ayton S, Frugier T, Steer D, et al. Type-1 interferons contribute to the neuroinflammatory response and disease progression of the MPTP mouse model of Parkinson’s disease. Glia. 2016;64:1590–604.
Abdullah A, Zhang M, Frugier T, Bedoui S, Taylor JM, Crack PJ. STING-mediated type-I interferons contribute to the neuroinflammatory process and detrimental effects following traumatic brain injury. J Neuroinflammation. 2018;15:323.
Karve IP, Zhang M, Habgood M, Frugier T, Brody KM, Sashindranath M, et al. Ablation of Type-1 IFN Signaling in Hematopoietic Cells Confers Protection Following Traumatic Brain Injury. eNeuro. 2016;3:ENEURO.0128–15.2016.
Teyra J, Singer AU, Schmitges FW, Jaynes P, Kit Leng Lui S, Polyak MJ, et al. Structural and functional characterization of ubiquitin variant inhibitors of USP15. Structure. 2019;27:590–605 e5.
Burkhart RA, Peng Y, Norris ZA, Tholey RM, Talbott VA, Liang Q, et al. Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol Cancer Res. 2013;11:901–11.
Gavory G, O’Dowd CR, Helm MD, Flasz J, Arkoudis E, Dossang A, et al. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat Chem Biol. 2018;14:118–25.
Anderson AC. The process of structure-based drug design. Chem Biol. 2003;10:787–97.
Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481–6.
Lamberto I, Liu X, Seo HS, Schauer NJ, Iacob RE, Hu W, et al. Structure-guided development of a potent and selective non-covalent active-site inhibitor of USP7. Cell Chem Biol. 2017;24:1490–1500 e11.
Funding
This work was supported by research award to P.G. from the Consortium Quebecois de la Recherche sur le Medicament, the Healthy Brains for Healthy Lives program at McGill University, Brain Canada, and Corbin Therapeutics, as well as a foundation research grant to P.G. from the Canadian Institutes of Health Research and from the Canadian Institute for Advanced Research (Human and the Microbiome Program). P.G. receives salary support from a Distinguished James McGill Professorship Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Georges, A., Gros, P. & Fodil, N. USP15: a review of its implication in immune and inflammatory processes and tumor progression. Genes Immun 22, 12–23 (2021). https://doi.org/10.1038/s41435-021-00125-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-021-00125-9
This article is cited by
-
RNA sequencing reveals differential long noncoding RNA expression profiles in bacterial and viral meningitis in children
BMC Medical Genomics (2024)
-
Targeting ubiquitin specific proteases (USPs) in cancer immunotherapy: from basic research to preclinical application
Journal of Experimental & Clinical Cancer Research (2023)
-
Genome-wide sequencing identifies a thermal-tolerance related synonymous mutation in the mussel, Mytilisepta virgata
Communications Biology (2023)
-
The Ubiquitin Proteasome System as a Therapeutic Area in Parkinson’s Disease
NeuroMolecular Medicine (2023)
-
Cell Biology of Parkin: Clues to the Development of New Therapeutics for Parkinson’s Disease
CNS Drugs (2022)