Abstract
UNC-93 homolog B1 (UNC93B1) is a key regulator of toll-like receptors (TLRs), pattern recognition receptors that sense invading pathogens and manage the innate immune response and deliver them from the endoplasmic reticulum to their respective endosomal signaling compartments. Several types of TLRs are known to contribute to the inflammatory process after allogeneic hematopoietic stem cell transplantation (SCT), so UNC93B1 might play integral roles there. We investigated the influence of the UNC93B1 single-nucleotide polymorphism (SNP) rs308328 (T>C) on transplant outcomes in a cohort of 237 patients undergoing unrelated HLA-matched bone marrow transplantation (BMT) for hematologic malignancies through the Japan Marrow Donor Program. The donor UNC93B1 C/C genotype was associated with a better 3-year overall survival than the donor UNC93B1 C/T or T/T genotype. An analysis of the UNC93B1 rs308328 genotype may therefore be useful for selecting the donor, estimating the prognosis, and creating therapeutic strategies after allogeneic SCT.
Similar content being viewed by others
Introduction
Allogeneic hematopoietic stem cell transplantation (SCT) is expected to cure hematologic malignancies. However, life-threatening complications associated with allogeneic SCT, such as severe infection, organ damage, and graft-versus-host disease (GVHD), remain obstacles to overcome [1]. Recently, increasing evidence has suggested that non-HLA genetic polymorphisms significantly influence outcomes after allogeneic SCT [2,3,4,5,6,7,8,9,10,11,12,13].
Toll-like receptors (TLRs) are the most important family of receptors in the early to middle stages of infectious immunity, cooperatively recognizing patterns present in microorganisms and augmenting the synthesis of inflammatory mediators [14,15,16,17]. Previous studies [18,19,20,21] have suggested that the TLR -signaling pathway plays important roles in the anti-microbial immunity and GVHD after allogeneic SCT. TLR genes have several functional single-nucleotide polymorphisms (SNPs), which have been shown to be associated with the survival outcomes after allogeneic SCT in our recent reports [22, 23]. TLRs are present not only on the cell surface but also inside the cell, and such intracellular TLRs are translocated from the ER to endolysosomes, which are critically regulated by UNC-93 homolog B1 (UNC93B1), an ER protein with 12 membrane-spanning domains [24]. UNC93B1 is encoded by the UNC93B1 gene on chromosome 11q13 and has one important SNP rs308328 (T>C) in an intronic region that is functional, and the major allele (T) has been reported to be associated with a lower UNC93B1 expression than T allele-negative individuals [25].
Given the above, we hypothesized that the SNP of UNC93B1 rs308328 may be associated with clinical outcomes after allogeneic SCT through affecting the function of UNC93B1.
Results
Frequencies of UNC93B1 genotypes
The rs308328 (T>C) polymorphism in the UNC93B1 gene was genotyped in 237 patients with hematologic malignancies and their unrelated donors in this cohort (Table 1). The frequencies of C/C, C/T, and T/T in the rs308328 (T>C) polymorphism were 14%, 40%, and 46% in the patients and 14%, 41%, and 46% in the donors (P = 0.98), respectively.
The prevalence of the UNC93B1 rs308328 C/C genotype in the healthy Japanese population was 17% in according to a database of 1000 Genomes (https://www.internationalgenome.org/home).
The UNC93B1 rs308328 genotype was also screened in 35 healthy Japanese volunteers in this study. The frequency of C/C in these volunteers was 11% (Table 1). The allele frequencies of this polymorphism in healthy controls, donors and patients did not differ to a statistically significant extent (P = 0.13).
Transplant outcomes according to the UNC93B1 rs308328 genotypes
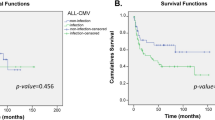
Univariate analyses (Table 2) showed that the donor UNC93B1 rs308328 C/C genotype was associated with a better 3-year overall survival (OS) than the donor UNC93B1 rs308328 C/T or T/T genotype (77% vs. 58%; P = 0.04; Fig. 1A). Three years was set as the study time point according to the median follow-up period among the survivors (753 days; range, 6–1918 days). The donor UNC93B1 rs308328 C/C genotype also exhibited a trend toward a lower 3-year transplant-related mortality (TRM; 13% vs. 27%, P = 0.06; Fig. 1C) than other genotypes but showed no marked reduction in the 3-year progression-free survival (PFS; 69% vs. 56%, P = 0.16; Fig. 1B) and 3-year relapse rate (18% vs. 18%, P = 0.72; Fig. 1D). The recipient UNC93B1 rs308328 genotype did not significantly influence the transplant outcomes in this study (Table 2).
The Kaplan–Meier analysis of the overall survival rates (A), the progression-free survival rates (B), the transplant-related mortality rates (C), and relapse rates (D) after transplantation according to the donor UNC93B1 rs308328 genotype. The solid lines represent the donor C/C genotype, and the dashed lines represent the donor T/T or C/T genotype.
After adjusting for clinical factors in the multivariate model (Table 3), the donor UNC93B1 rs308328 C/C genotype remained associated with a better 3-year OS than other genotypes (hazard ratio [HR], 0.37; 95% confidence interval [CI], 0.16–0.88; P = 0.03).
When the main causes of death were analyzed according to the UNC93B1 rs308328 genotype, the donor UNC93B1 rs308328 C/C genotype showed half the incidence of death attributed to infection compared with other genotypes (Fig. 2), although there were no significant differences.
Discussion
The present study showed that the donor UNC93B1 rs308328 C/C genotype, which is presumed to have greater inducibility of UNC93B1 than other genotypes [25], was associated with significantly better survival outcomes than other genotypes in patients with hematological malignancies receiving HLA fully matched unrelated bone marrow transplantation (BMT).
The mechanisms through which the donor UNC93B1 rs308328 C/C genotype exerts its beneficial effects remain to be determined. One plausible explanation is that patients transplanted from donors with the UNC93B1 C/C genotype may have been less susceptible to infection than those without the UNC93B1 C/C genotype, considering that the incidence of death mainly attributed to infections was 3.0% vs. 8.3%, respectively (Fig. 2A). Evidence supporting this hypothesis may be seen in a previous report [26], in which two unrelated children possessing autosomal recessive deficiency of UNC93B1 developed encephalitis due to herpes-simplex virus, wherein the antiviral cellular responses were impaired in the interferon-α (IFN-α), IFN-β, and IFN-λ pathways. Experimental studies [26,27,28,29,30,31,32], have shown that a lack or suppression of UNC93B1 inhibits the activation of TLR3, TLR7, and TLR9 to induce inflammatory mediators, leading to increased susceptibility to infections. In addition, mouse model studies using UNC93B1 mutant mice, which lack the UNC93B1 function as a result of H412R missense mutation, exhibited increased susceptibility to Toxoplasma gondii [27], Trypanosoma cruzi [28], Leishmania major [29], and cytomegalovirus (CMV) [31] infections in association with the reduced expression of inflammatory mediators, including IFN-γ (T. cruzi, CMV, L. major), IFN-α (CMV) and interleukin (IL)-12p40 (T. gondii, T. cruzi). In another experimental study [32], other UNC93B1 mutant mice with a 54-amino acid deletion in exon 4, which are also deficient in functional UNC93B1, showed a decreased number of activated exudate macrophages and the decreased expression of CXC Chemokine Ligand (CXCL) 10, IFN-γ, and type I IFN in the early phase of influenza A H1N1 infection. Given these previous findings, it is reasonable to assume from the present study that the donor UNC93B1 C/C genotype, potentially having higher inducibility of UNC93B1 than other genotypes, may enhance the functions of TLRs, thereby augmenting infectious immunity, which improved the survival outcomes after allogeneic SCT.
There is a contrasting report that the UNC93B1 mutation with a 54-amino acid deletion in exon 4 was associated with the increased cardiac expression of IFN-β and markers of tissue injury and fibrosis early after coxsackievirus strain B serotype 3 (CVB3), which is a picornavirus that induces myocarditis [33]. However, one or more alternative pathways may mediate the cardiac IFN-β upregulation in the case of CSV3. For example, melanoma differentiation-associated gene 5 (MDA5) detects the double-stranded RNA replicative form in picornavirus-infected cells and induces the production of inflammatory cytokines, including IFN-β [30]. Since different immune system cascades are activated depending on the type of pathogen, another cohort study to validate the current findings is desired in order to confirm the impact of UNC93B1 on allogeneic SCT, considering the type of pathogen, as this is beyond the scope of the present study.
Some reports have suggested that TLR4 polymorphisms may have an impact directly on the infection and graft rejection after transplantation [34,35,36,37,38]. We showed that the donor TLR4 + 3725G/G genotype, which induces a low expression of TLR4, was significantly associated with a lower incidence of fatal infections than the donor G/C and C/C genotypes in another cohort of 367 patients who underwent unrelated HLA-matched BMT for hematologic malignancies through the Japan Marrow Donor Program (JMDP) [22]. The UNC93B1 gene and the TLR4 gene are not predicted to be in linkage disequilibrium because the UNC93B1 gene is located on chromosome 11q13, while the TLR4 gene is located on chromosome 9q32-q33. Furthermore, the UNC93B1 gene does not affect the function of TLR directly. Therefore, it is reasonable to consider that the donor UNC93B1 rs308328 C/C genotype, which putatively has higher inducibility of UNC93B1 than others, improves the outcomes of transplantation without the effects of TLR4 SNPs.
Several limitations associated with the present study warrant mention. First, detailed information on the infections, including the types, severity, treatments, and therapeutic appropriateness, was not obtained in the current study. Second, the functional roles of the SNP of UNC93B1 rs308328 in BMT remain purely speculative due to the lack of data using blood samples from BMT recipients and their donors. Further studies including more samples from patients, donors, and healthy individuals need to be conducted.
Another limitation is that our results for the UNC93B1 gene may be a coincidence in the analysis of polymorphisms in many genes associated with the UNC93B1 gene, especially other TLR-related genes, although the fact that UNC93B1 is an important protein for infectious immunity that integrates the functions of TLRs and that the UNC93B1 gene plays a functional role suggests that the UNC93B1 gene polymorphism is probably responsible for the observed phenotype. Therefore, further studies using a separate cohort should be warranted to clarify whether polymorphisms in TLR-related genes are involved in the outcomes of BMT in an integrated manner.
In conclusion, the findings of the present study suggested that the donor UNC93B1 rs308328 C/C genotype predicted better survival outcomes after SCT than other genotypes. Therefore, the donor UNC93B1 rs308328 C/C genotype in donors may be a valuable tool for selecting donors and evaluating pretransplantation risks that, combined with other currently known risk factors, can form the basis for carrying out suitable tailoring of transplantation strategies. Considering the plausible functional roles of these polymorphisms, they may be candidates for future prophylactic and therapeutic strategies for complications after allogeneic SCT and may lead to the development of molecular targeted therapy [39]. Further studies are warranted to ascertain whether or not the findings of this study can be extended to other stem cell sources or to HLA-mismatched transplantation and to validate the present findings in other ethnic groups.
Materials and methods
Patients
A total of 237 patients and their unrelated donors were included in the study. The patients underwent HLA-matched BMT for hematologic malignancies through the JMDP with T cell-replete marrow from HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 allele-matched donors between May 2006 and April 2009. No patient had a history of any previous transplantation. The final clinical data analyses of these patients were completed by September 27, 2011.
The diagnosed diseases were acute myeloid leukemia (AML) (n = 115; 49%), acute lymphoblastic leukemia (ALL) (n = 46; 19%), myelodysplastic syndrome (MDS) (n = 38; 16%), malignant lymphoma (ML) (n = 24; 10%), chronic myeloid leukemia (CML) (n = 10; 4%), myeloproliferative neoplasm (MPN) (n = 3; 1%), and multiple myeloma (MM) (n = 1; 0.4%) (Table 1). Relapse or secondary cases were defined as high-risk diseases. Myeloid malignancies included AML, MDS, CML, and MPD. Lymphoid malignancies included ALL, ML, and MM.
The conditioning regimen depends on the type of underlying disease and the patient’s condition. The combination of cyclophosphamide (CY) and total body irradiation (TBI) was mainly classified as myeloablative conditioning (MAC), whereas the combination of fludarabine and melphalan was mainly classified as reduced-intensity conditioning (RIC) [40]. Cyclosporin- or tacrolimus-based therapy was selected as GVHD prophylaxis [41, 42]. Patients who were used anti-T cell therapy, such as anti-thymocyte globulin or ex vivo T cell depletion were excluded from this study.
All patients and donors provided their informed consent at the time of transplantation to participate in molecular studies of this nature according to the Declaration of Helsinki. This project was approved by the Institutional Review Board of Aichi Medical University School of Medicine and the JMDP.
UNC93B1 genotyping
Real-time polymerase chain reaction (PCR) genotyping for UNC93B1 was done with the TaqMan-Allelic discrimination method in a Step One Plus Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) as described previously [7], and the results were analyzed using the Allelic Discrimination software program (Applied Biosystems). We purchased the specific probe designed for SNP rs308328 (T>C) (product No. C_2623961_1) and TaqMan genotyping master mix from Applied Biosystems.
Data management and statistical analyses
The JMDP collected the data using a standardized report form. Follow-up reports were submitted at 100 days, 1 year, and annually after transplantation. Pre-transplant cytomegalovirus (CMV) serostatus was routinely measured only in patients but not in their donors. Neutrophil engraftment was confirmed by an absolute neutrophil count of more than 0.5 × 109/L for at least three consecutive days. Acute GVHD (aGVHD) was diagnosed and graded in accordance with the established criteria [43]. Chronic GVHD (cGVHD) was classified on the basis of the Seattle criteria [44].
We calculated the OS from the date of transplantation to death, regardless of cause, and defined disease relapse as the number of days from transplantation to disease relapse or progression. The TRM was defined as death due to any cause without relapse or disease progression, including infections, toxicities, and other non-relapse- or disease progression-related causes of death. The PFS was defined as the survival without disease relapse or progression. Any patients who were alive at the last follow-up date were censored. Data concerning the clinical and microbiologic features of infections, postmortem changes, prophylaxis of infections, and therapy of GVHD given on an institutional basis were not available for this study.
The EZR software package [45] was used in all statistical analyses. The probabilities of the OS and PFS were calculated with the Kaplan–Meier method, and comparisons between groups were made with the log-rank test. The occurrence of TRM, disease relapse, aGVHD, and cGVHD was compared using the Gray test [46] and analyzed using a cumulative incidence analysis [47], considering relapse, death without disease relapse, death without aGVHD, death without cGVHD, and death without engraftment as respective competing risks. A multivariate Cox model was created for the OS, TRM, relapse, grades II–IV aGVHD, grades III–IV aGVHD, and cGVHD, using stepwise selection at a significance level of 5% to evaluate the hazard ratio (HR) associated with the UNC93B1 rs308328 genotype. The recipient age at the time of BMT, sex, pretransplantation CMV serostatus, performance status, disease characteristics (i.e., disease type, disease lineage, and disease risk at transplantation), donor characteristics (i.e., age, sex, sex compatibility, and ABO compatibility), transplant characteristics (i.e., MAC or RIC and total nucleated cell count harvested per recipient weight), and year of transplantation were included as variables. The median was used as the cut-off point for continuous variables.
The chi-square test and Mann–Whitney test were used to compare the results of the two groups. The Hardy–Weinberg equilibrium for the UNC93B1 rs308328 gene polymorphism was tested using the Haploview program. For both the univariate and multivariate analyses P < 0.05 was considered to indicate statistical significance.
References
Gratwohl A, Brand R, Frassoni F, Rocha V, Niederwieser D, Reusser P, et al. Cause of death after allogeneic haematopoietic stem cell transplantation (HSCT) in early leukaemias: an EBMT analysis of lethal infectious complications and changes over calendar time. Bone Marrow Transpl. 2005;36:757–69.
Nomoto H, Takami A, Espinoza JL, Onizuka M, Kashiwase K, Morishima Y, et al. Recipient ADAMTS13 single-nucleotide polymorphism predicts relapse after unrelated bone marrow transplantation for hematologic malignancy. Int J Mol Sci. 2019;20:214.
Takami A, Espinoza JL, Onizuka M, Ishiyama K, Kawase T, Kanda Y, et al. A single-nucleotide polymorphism of the Fcgamma receptor type IIIA gene in the recipient predicts transplant outcomes after HLA fully matched unrelated BMT for myeloid malignancies. Bone Marrow Transpl. 2011;46:238–43.
Espinoza JL, Takami A, Onizuka M, Morishima Y, Fukuda T, Kodera Y, et al. Recipient PTPN22 -1123 C/C genotype predicts acute graft-versus-host disease after HLA fully matched unrelated bone marrow transplantation for hematologic malignancies. Biol Blood Marrow Transplant. 2013;19:240–6.
Nakata K, Takami A, Espinoza JL, Matsuo K, Morishima Y, Onizuka M, et al. The recipient CXCL10 + 1642C>G variation predicts survival outcomes after HLA fully matched unrelated bone marrow transplantation. Clin Immunol. 2013;146:104–11.
Horio T, Mizuno S, Uchino K, Mizutani M, Hanamura I, Espinoza JL, et al. The recipient CCR5 variation predicts survival outcomes after bone marrow transplantation. Transpl Immunol. 2017;42:34–9.
Espinoza JL, Takami A, Onizuka M, Sao H, Akiyama H, Miyamura K, et al. NKG2D gene polymorphism has a significant impact on transplant outcomes after HLA-fully-matched unrelated bone marrow transplantation for standard risk hematologic malignancies. Haematologica. 2009;94:1427–34.
Espinoza LJ, Takami A, Nakata K, Yamada K, Onizuka M, Kawase T, et al. Genetic variants of human granzyme B predict transplant outcomes after HLA matched unrelated bone marrow transplantation for myeloid malignancies. PLoS ONE. 2011;6:e23827.
Espinoza JL, Takami A, Nakata K, Onizuka M, Kawase T, Akiyama H, et al. A genetic variant in the IL-17 promoter is functionally associated with acute graft-versus-host disease after unrelated bone marrow transplantation. PLoS ONE. 2011;6:e26229.
Nomoto H, Takami A, Espinoza JL, Matsuo K, Mizuno S, Onizuka M, et al. A donor thrombomodulin gene variation predicts graft-versus-host disease development and mortality after bone marrow transplantation. Int J Hematol. 2015;102:460–70.
Suetsugu K, Mori Y, Yamamoto N, Shigematsu T, Miyamoto T, Egashira N, et al. Impact of CYP3A5, POR, and CYP2C19 polymorphisms on trough concentration to dose ratio of tacrolimus in allogeneic hematopoietic stem cell transplantation. Int J Mol Sci. 2019;20:2413.
Koldej RM, Perera T, Collins J, Ritchie DS. Association between P2X7 polymorphisms and post-transplant outcomes in allogeneic haematopoietic stem cell transplantation. Int J Mol Sci. 2020;21:3772.
Horio T, Morishita E, Mizuno S, Uchino K, Hanamura I, Espinoza JL, et al. Donor heme oxygenase-1 promoter gene polymorphism predicts survival after unrelated bone marrow transplantation for high-risk patients. Cancers 2020;12:424.
Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20.
Schnetzke U, Spies-Weisshart B, Yomade O, Fischer M, Rachow T, Schrenk K, et al. Polymorphisms of Toll-like receptors (TLR2 and TLR4) are associated with the risk of infectious complications in acute myeloid leukemia. Genes Immun. 2015;16:83–8.
Trejo-de la OA, Hernandez-Sancen P, Maldonado-Bernal C. Relevance of single-nucleotide polymorphisms in human TLR genes to infectious and inflammatory diseases and cancer. Genes Immun. 2014;15:199–209.
Shey MS, Randhawa AK, Bowmaker M, Smith E, Scriba TJ, de Kock M, et al. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin-6 secretion. Genes Immun. 2010;11:561–72.
Heidegger S, van den Brink MR, Haas T, Poeck H. The role of pattern-recognition receptors in graft-versus-host disease and graft-versus-leukemia after allogeneic stem cell transplantation. Front Immunol. 2014;5:337.
Penack O, Holler E, van den Brink MR. Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115:1865–72.
Maeda Y. Pathogenesis of graft-versus-host disease: innate immunity amplifying acute alloimmune responses. Int J Hematol. 2013;98:293–9.
Tu S, Zhong D, Xie W, Huang W, Jiang Y, Li Y. Role of toll-like receptor signaling in the pathogenesis of graft-versus-host diseases. Int J Mol Sci. 2016;17:1288.
Uchino K, Mizuno S, Sato-Otsubo A, Nannya Y, Mizutani M, Horio T, et al. Toll-like receptor genetic variations in bone marrow transplantation. Oncotarget. 2017;8:45670–86.
Uchino K, Mizuno S, Mizutani M, Horio T, Hanamura I, Espinoza JL, et al. Toll-like receptor 1 variation increases the risk of transplant-related mortality in hematologic malignancies. Transpl Immunol. 2016;38:60–6.
Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–8.
Inoue N, Katsumata Y, Watanabe M, Ishido N, Manabe Y, Watanabe A, et al. Polymorphisms and expression of toll-like receptors in autoimmune thyroid diseases. Autoimmunity. 2017;50:182–91.
Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–12.
Melo MB, Kasperkovitz P, Cerny A, Konen-Waisman S, Kurt-Jones EA, Lien E, et al. UNC93B1 mediates host resistance to infection with Toxoplasma gondii. PLoS Pathog. 2010;6:e1001071.
Caetano BC, Carmo BB, Melo MB, Cerny A, dos Santos SL, Bartholomeu DC, et al. Requirement of UNC93B1 reveals a critical role for TLR7 in host resistance to primary infection with Trypanosoma cruzi. J Immunol. 2011;187:1903–11.
Schamber-Reis BLF, Petritus PM, Caetano BC, Martinez ER, Okuda K, Golenbock D, et al. UNC93B1 and nucleic acid-sensing toll-like receptors mediate host resistance to infection with Leishmania major. J Biol Chem. 2013;288:7127–36.
Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5.
Crane MJ, Gaddi PJ, Salazar-Mather TP. UNC93B1 mediates innate inflammation and antiviral defense in the liver during acute murine cytomegalovirus infection. PLoS ONE. 2012;7:e39161.
Lafferty EI, Flaczyk A, Angers I, Homer R, d’Hennezel E, Malo D, et al. An ENU-induced splicing mutation reveals a role for Unc93b1 in early immune cell activation following influenza A H1N1 infection. Genes Immun. 2014;15:320–32.
Lafferty EI, Wiltshire SA, Angers I, Vidal SM, Qureshi ST. Unc93b1-dependent endosomal toll-like receptor signaling regulates inflammation and mortality during coxsackievirus B3 infection. J Innate Immun. 2015;7:315–30.
Noreen M, Shah MA, Mall SM, Choudhary S, Hussain T, Ahmed I, et al. TLR4 polymorphisms and disease susceptibility. Inflamm Res. 2012;61:177–88.
Oetting WS, Guan W, Schladt DP, Leduc RE, Jacobson PA, Matas AJ, et al. Donor polymorphisms of toll-like receptor 4 associated with graft failure in liver transplant recipients. Liver Transpl. 2012;18:1399–405.
Lee SO, Brown RA, Kang SH, Abdel Massih RC, Razonable RR. Toll-like receptor 4 polymorphisms and the risk of gram-negative bacterial infections after liver transplantation. Transplantation. 2011;92:690–6.
Dhillon N, Walsh L, Kruger B, Ward SC, Godbold JH, Radwan M, et al. A single nucleotide polymorphism of Toll-like receptor 4 identifies the risk of developing graft failure after liver transplantation. J Hepatol. 2010;53:67–72.
Hwang YH, Ro H, Choi I, Kim H, Oh KH, Hwang JI, et al. Impact of polymorphisms of TLR4/CD14 and TLR3 on acute rejection in kidney transplantation. Transplantation. 2009;88:699–705.
Raschella G, Melino G, Gambacurta A. Cell death in cancer in the era of precision medicine. Genes Immun. 2019;20:529–38.
Giralt S, Ballen K, Rizzo D, Bacigalupo A, Horowitz M, Pasquini M, et al. Reduced-intensity conditioning regimen workshop: defining the dose spectrum. Report of a workshop convened by the center for international blood and marrow transplant research. Biol Blood Marrow Transplant. 2009;15:367–9.
Nash RA, Antin JH, Karanes C, Fay JW, Avalos BR, Yeager AM, et al. Phase 3 study comparing methotrexate and tacrolimus with methotrexate and cyclosporine for prophylaxis of acute graft-versus-host disease after marrow transplantation from unrelated donors. Blood. 2000;96:2062–8.
Storb R, Deeg HJ, Whitehead J, Appelbaum F, Beatty P, Bensinger W, et al. Methotrexate and cyclosporine compared with cyclosporine alone for prophylaxis of acute graft versus host disease after marrow transplantation for leukemia. N Engl J Med. 1986;314:729–35.
Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, et al. 1994 Consensus conference on acute GVHD grading. Bone Marrow Transpl. 1995;15:825–8.
Shulman HM, Sullivan KM, Weiden PL, McDonald GB, Striker GE, Sale GE, et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am J Med. 1980;69:204–17.
Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013;48:452–8.
Gooley TA, Leisenring W, Crowley J, Storer BE. Estimation of failure probabilities in the presence of competing risks: new representations of old estimators. Stat Med. 1999;18:695–706.
Scrucca L, Santucci A, Aversa F. Competing risk analysis using R: an easy guide for clinicians. Bone Marrow Transpl. 2007;40:381–7.
Acknowledgements
This study was supported by grants from the Ministry of Education, Culture, Sports and Technology of Japan, a Research on Allergic Disease and Immunology (H26-106) in Health and Labor Science Grant from the Ministry of Health, Labour and Welfare of Japan, the SENSHIN Medical Research Foundation (Osaka, Japan), the Aichi Cancer Research Foundation (Nagoya, Japan), and the 24th General Assembly of the Japanese Association of Medical Sciences (Nagoya, Japan). The funders played no role in the study design, data collection, and analysis, the decision to publish, or the preparation of the manuscript. We thank all of the Japan Marrow Donor Program (JMDP) transplant teams who provided valuable assistance in caring for the patients and donors evaluated in this study. assistance in caring for the patients and donors investigated in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Uchino, K., Vu Quang, L., Mizuno, S. et al. Donor UNC-93 Homolog B1 genetic polymorphism predicts survival outcomes after unrelated bone marrow transplantation. Genes Immun 22, 35–43 (2021). https://doi.org/10.1038/s41435-021-00122-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-021-00122-y