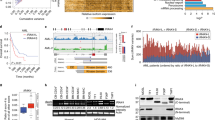
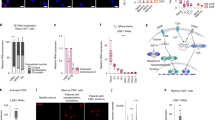
Abstract
Human IL12RB1 is an autosomal gene that is essential for mycobacterial disease resistance and T cell differentiation. Using primary human tissue and PBMCs, we demonstrate that lung and T cell IL12RB1 expression is allele-biased, and the extent to which cells express one IL12RB1 allele is unaffected by activation. Furthermore following its expression the IL12RB1 pre-mRNA is processed into either IL12RB1 Isoform 1 (IL12Rβ1, a positive regulator of IL12 responsiveness) or IL12RB1 Isoform 2 (a protein of heretofore unknown function). T cells choice to process pre-mRNA into Isoform 1 or Isoform 2 is controlled by intragenic competition of IL12RB1 exon 9–10 splicing with IL12RB1 exon 9b splicing, as well as an IL12RB1 exon 9b-associated polyadenylation site. Heterogeneous nuclear ribonucleoprotein H (hnRNP H) binds near the regulated polyadenylation site, but is not required for exon 9b polyadenylation. Finally, microRNA-mediated knockdown experiments demonstrated that IL12RB1 Isoform 2 promotes T cell IL12 responses. Collectively, our data support a model wherein tissue expression of human IL12RB1 is allele-biased and produces an hnRNP H-bound pre-mRNA, the processing of which generates a novel IL12 response regulator.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin Immunol. 2014;26:454–70.
de Beaucoudrey L, Samarina A, Bustamante J, Cobat A, Boisson-Dupuis S, Feinberg J, et al. Revisiting human IL-12Rbeta1 deficiency: a survey of 141 patients from 30 countries. Med (Baltim). 2010;89:381–402.
Robinson RT. IL12Rbeta1: the cytokine receptor that we used to know. Cytokine. 2015;71:348–59.
van de Vosse E, Haverkamp MH, Ramirez-Alejo N, Martinez-Gallo M, Blancas-Galicia L, Metin A, et al. IL-12Rbeta1 deficiency: mutation update and description of the IL12RB1 variation database. Hum Mutat. 2013;34:1329–39.
Dhiman N, Ovsyannikova IG, Cunningham JM, Vierkant RA, Kennedy RB, Pankratz VS, et al. Associations between measles vaccine immunity and single-nucleotide polymorphisms in cytokine and cytokine receptor genes. J Infect Dis. 2007;195:21–9.
White SJ, Haralambieva IH, Ovsyannikova IG, Vierkant RA, O’Byrne MM, Poland GA. Replication of associations between cytokine and cytokine receptor single nucleotide polymorphisms and measles-specific adaptive immunophenotypic extremes. Hum Immunol. 2012;73:636–40.
Li X, Hawkins GA, Ampleford EJ, Moore WC, Li H, Hastie AT, et al. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J Allergy Clin Immunol. 2013;132:313–20. e15
Takahashi N, Akahoshi M, Matsuda A, Ebe K, Inomata N, Obara K, et al. Association of the IL12RB1 promoter polymorphisms with increased risk of atopic dermatitis and other allergic phenotypes. Hum Mol Genet. 2005;14:3149–59.
Hong X, Wang G, Liu X, Kumar R, Tsai HJ, Arguelles L, et al. Gene polymorphisms, breast-feeding, and development of food sensitization in early childhood. J Allergy Clin Immunol. 2011;128:374–81. e2
Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25.
Presky DH, Yang H, Minetti LJ, Chua AO, Nabavi N, Wu CY, et al. A functional interleukin 12 receptor complex is composed of two beta-type cytokine receptor subunits. Proc Natl Acad Sci USA. 1996;93:14002–7.
Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–708.
Khader SA, Pearl JE, Sakamoto K, Gilmartin L, Bell GK, Jelley-Gibbs DM, et al. IL-23 compensates for the absence of IL-12p70 and is essential for the IL-17 response during tuberculosis but is dispensable for protection and antigen-specific IFN-gamma responses if IL-12p70 is available. J Immunol. 2005;175:788–95.
Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–7.
Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–54.
Ford NR, Miller HE, Reeme AE, Waukau J, Bengtson C, Routes JM, et al. Inflammatory signals direct expression of human IL12RB1 into multiple distinct isoforms. J Immunol. 2012;189:4684–94.
Ray AA, Fountain JJ, Miller HE, Cooper AM, Robinson RT. IL12Rbeta1DeltaTM is a secreted product of il12rb1 that promotes control of extrapulmonary tuberculosis. Infect Immun. 2015;83:560–71.
Reinius B, Sandberg R. Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat Rev Genet. 2015;16:653–64.
Chang X. RNA-binding protein hnRNPLL as a critical regulator of lymphocyte homeostasis and differentiation. Wiley Interdiscip Rev RNA. 2016;7:295–302.
Martinez NM, Lynch KW. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol Rev. 2013;253:216–36.
Ku CJ, Lim KC, Kalantry S, Maillard I, Engel JD, Hosoya T. A monoallelic-to-biallelic T-cell transcriptional switch regulates GATA3 abundance. Genes Dev. 2015;29:1930–41.
Brady BL, Steinel NC, Bassing CH. Antigen receptor allelic exclusion: an update and reappraisal. J Immunol. 2010;185:3801–8.
Bayley JP, van Rietschoten JG, Bakker AM, van Baarsen L, Kaijzel EL, Wierenga EA, et al. Allele-specific expression of the IL-1 alpha gene in human CD4+T cell clones. J Immunol. 2003;171:2349–53.
Bix M, Locksley RM. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+T cells. Science. 1998;281:1352–4.
Calado DP, Paixao T, Holmberg D, Haury M. Stochastic monoallelic expression of IL-10 in T cells. J Immunol. 2006;177:5358–64.
Hollander GA, Zuklys S, Morel C, Mizoguchi E, Mobisson K, Simpson S, et al. Monoallelic expression of the interleukin-2 locus. Science. 1998;279:2118–21.
Kelly BL, Locksley RM. Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J Immunol. 2000;165:2982–6.
Rhoades KL, Singh N, Simon I, Glidden B, Cedar H, Chess A. Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr Biol. 2000;10:789–92.
Riviere I, Sunshine MJ, Littman DR. Regulation of IL-4 expression by activation of individual alleles. Immunity. 1998;9:217–28.
van Rietschoten JG, Verzijlbergen KF, Gringhuis SI, van der Pouw Kraan TC, Bayley JP, Wierenga EA, et al. Differentially methylated alleles in a distinct region of the human interleukin-1alpha promoter are associated with allele-specific expression of IL-1alpha in CD4+T cells. Blood. 2006;108:2143–9.
Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1’s multifunctional regulatory roles. RNA. 2010;16:1449–62.
Yeh YM, Chen CY, Huang PR, Hsu CW, Wu CC, Wang TC. Proteomic analyses of genes regulated by heterogeneous nuclear ribonucleoproteins A/B in Jurkat cells. Proteomics. 2014;14:1357–66.
Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol. 2001;21:1228–38.
Gaudreau MC, Heyd F, Bastien R, Wilhelm B, Moroy T. Alternative splicing controlled by heterogeneous nuclear ribonucleoprotein L regulates development, proliferation, and migration of thymic pre-T cells. J Immunol. 2012;188:5377–88.
Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–802.
Shankarling G, Cole BS, Mallory MJ, Lynch KW. Transcriptome-wide RNA interaction profiling reveals physical and functional targets of hnRNP L in human T cells. Mol Cell Biol. 2014;34:71–83.
Tong A, Nguyen J, Lynch KW. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J Biol Chem. 2005;280:38297–304.
Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA. 2008;14:2038–49.
Chang X, Li B, Rao A. RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation. Proc Natl Acad Sci USA. 2015;112:E1888–97.
Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–91.
Wu Z, Jia X, de la Cruz L, Su XC, Marzolf B, Troisch P, et al. Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity. 2008;29:863–75.
Meininger I, Griesbach RA, Hu D, Gehring T, Seeholzer T, Bertossi A, et al. Alternative splicing of MALT1 controls signalling and activation of CD4(+) T cells. Nat Commun. 2016;7:11292.
Turner AJ, Aggarwal P, Miller HE, Waukau J, Routes JM, Broeckel U, et al. The introduction of RNA-DNA differences underlies interindividual variation in the human IL12RB1 mRNA repertoire. Proc Natl Acad Sci USA. 2015;112:15414–9.
Miller HE, Robinson RT. Early control of Mycobacterium tuberculosis infection requires il12rb1 expression by rag1-dependent lineages. Infect Immun. 2012;80:3828–41.
Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF. Programming for CD8 T cell memory development requires IL-12 or type I IFN. J Immunol. 2009;182:2786–94.
van de Vosse E, de Paus RA, van Dissel JT, Ottenhoff TH. Molecular complementation of IL-12Rbeta1 deficiency reveals functional differences between IL-12Rbeta1 alleles including partial IL-12Rbeta1 deficiency. Hum Mol Genet. 2005;14:3847–55.
Abraham RT, Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat Rev Immunol. 2004;4:301–8.
Lou H, Cote GJ, Gagel RF. The calcitonin exon and its flanking intronic sequences are sufficient for the regulation of human calcitonin/calcitonin gene-related peptide alternative RNA splicing. Mol Endocrinol. 1994;8:1618–26.
Lou H, Gagel RF. Alternative RNA processing--its role in regulating expression of calcitonin/calcitonin gene-related peptide. J Endocrinol. 1998;156:401–5.
Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3’ end processing regulation. Nucleic Acids Res. 2010;38:2757–74.
Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H’/F/2H9 family. J Biol Chem. 2001;276:43850–9.
Fogel BL, McNally MT. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J Biol Chem. 2000;275:32371–8.
Hastings ML, Wilson CM, Munroe SH. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA. 2001;7:859–74.
Kralovicova J, Vorechovsky I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J Immunol. 2006;176:2381–8.
Marcucci R, Baralle FE, Romano M. Complex splicing control of the human Thrombopoietin gene by intronic G runs. Nucleic Acids Res. 2007;35:132–42.
Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol. 2008;28:5403–19.
McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–71.
McNally LM, Yee L, McNally MT. Heterogeneous nuclear ribonucleoprotein H is required for optimal U11 small nuclear ribonucleoprotein binding to a retroviral RNA-processing control element: implications for U12-dependent RNA splicing. J Biol Chem. 2006;281:2478–88.
Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci USA. 2004;101:15700–5.
Dominguez C, Allain FH. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–45.
Fieschi C, Dupuis S, Catherinot E, Feinberg J, Bustamante J, Breiman A, et al. Low penetrance, broad resistance, and favorable outcome of interleukin 12 receptor beta1 deficiency: medical and immunological implications. J Exp Med. 2003;197:527–35.
Chua AO, Wilkinson VL, Presky DH, Gubler U. Cloning and characterization of a mouse IL-12 receptor-beta component. J Immunol. 1995;155:4286–94.
Robinson RT, Khader SA, Martino CA, Fountain JJ, Teixeira-Coelho M, Pearl JE, et al. Mycobacterium tuberculosis infection induces il12rb1 splicing to generate a novel IL-12Rbeta1 isoform that enhances DC migration. J Exp Med. 2010;207:591–605.
Showe LC, Wysocka M, Wang B, Lineman-Williams D, Peritt D, Showe MK, et al. Structure of the mouse IL-12R beta 1 chain and regulation of its expression in BCG/LPS-treated mice. Ann N Y Acad Sci. 1996;795:413–5.
Yamamoto K, Kobayashi H, Miura O, Hirosawa S, Miyasaka N. Assignment of IL12RB1 and IL12RB2, interleukin-12 receptor beta 1 and beta 2 chains, to human chromosome 19 bandp13.1 and chromosome 1 band p31.2, respectively, by in situ hybridization. Cytogenet Cell Genet. 1997;77:257–8.
Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
Akahoshi M, Nakashima H, Miyake K, Inoue Y, Shimizu S, Tanaka Y, et al. Influence of interleukin-12 receptor beta1 polymorphisms on tuberculosis. Hum Genet. 2003;112:237–43.
Hussain SK, Madeleine MM, Johnson LG, Du Q, Galloway DA, Daling JR, et al. Nucleotide variation in IL-10 and IL-12 and their receptors and cervical and vulvar cancer risk: a hybrid case-parent triad and case-control study. Int J Cancer. 2013;133:201–13.
Quan L, Gong Z, Yao S, Bandera EV, Zirpoli G, Hwang H, et al. Cytokine and cytokine receptor genes of the adaptive immune response are differentially associated with breast cancer risk in American women of African and European ancestry. Int J Cancer. 2014;134:1408–21.
Giatrakos S, Huse K, Kanni T, Tzanetakou V, Kramer M, Grech I, et al. Haplotypes of IL-12Rbeta1 impact on the clinical phenotype of hidradenitis suppurativa. Cytokine. 2013;62:297–301.
Esteves LM, Bulhoes SM, Branco CC, Mota FM, Paiva C, Cabral R, et al. Human leptospirosis: seroreactivity and genetic susceptibility in the population of Sao Miguel Island (Azores, Portugal). PLoS One. 2014;9:e108534.
Ruhrmann S, Stridh P, Kular L, Jagodic M. Genomic imprinting: a missing piece of the Multiple Sclerosis puzzle? Int J Biochem Cell Biol. 2015;67:49–57.
Monahan K, Lomvardas S. Monoallelic expression of olfactory receptors. Annu Rev Cell Dev Biol. 2015;31:721–40.
Kim J, Bartel DP. Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nat Biotechnol. 2009;27:472–7.
Miller CL, Haas U, Diaz R, Leeper NJ, Kundu RK, Patlolla B, et al. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014;10:e1004263.
Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–98.
Gautrey H, Jackson C, Dittrich AL, Browell D, Lennard T, Tyson-Capper A. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 2015;12:1139–51.
Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–86.
McNally MT. RNA processing control in avian retroviruses. Front Biosci. 2008;13:3869–83.
Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–67.
Uren PJ, Bahrami-Samani E, de Araujo PR, Vogel C, Qiao M, Burns SC, et al. High-throughput analyses of hnRNP H1 dissects its multi-functional aspect. RNA Biol. 2016;13:400–11.
Gendrel AV, Attia M, Chen CJ, Diabangouaya P, Servant N, Barillot E, et al. Developmental dynamics and disease potential of random monoallelic gene expression. Dev Cell. 2014;28:366–80.
Denning W, Das S, Guo S, Xu J, Kappes JC, Hel Z. Optimization of the transductional efficiency of lentiviral vectors: effect of sera and polycations. Mol Biotechnol. 2013;53:308–14.
Acknowledgements
We would like to thank and acknowledge the following individuals, whose work contributed to indicated Figures: Jill Waukau and Christine Bengtson (Figs. 1 and 2), Mark McNally and Lisa McNally (Figs. 4 and 5), Katrina Monson and Brady Brooks (Fig. 6). We would also like to thank Abigail Robinson for the chromosome 19 karyotype image in Fig. 7. This work was supported by the Medical College of Wisconsin (MCW) and National Institutes of Health grant R01 AI121212 (to RTR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Reeme, A.E., Claeys, T.A., Aggarwal, P. et al. Human IL12RB1 expression is allele-biased and produces a novel IL12 response regulator. Genes Immun 20, 181–197 (2019). https://doi.org/10.1038/s41435-018-0023-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-018-0023-2