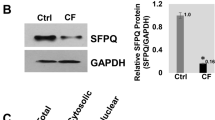
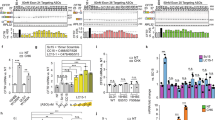
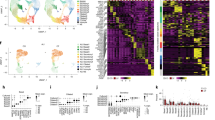
Abstract
Transcription of the cystic fibrosis transmembrane conductance regulator (CFTR) gene is regulated by both ubiquitous and cell-type selective cis-regulatory elements (CREs). These CREs include extragenic and intronic enhancers that bind lineage-specific transcription factors, and architectural protein-marked structural elements. Deletion of the airway-selective -35 kb enhancer in 16HBE14o− lung epithelial cells was shown earlier to disrupt normal enhancer-promoter looping at the CFTR locus and nearly abolish its expression. Using a 16HBE14o− clone that lacks the endogenous -35 kb CRE, we explore the impact of relocating the functional core of this element to an ectopic site in intron 1. The -35 kb sequence establishes a de novo enhancer signature in chromatin at the insertion site, and augments CFTR expression, albeit not fully restoring WT levels. The relocated -35 kb enhancer also initiates de novo chromatin contacts with the CFTR promoter and other known CFTR CREs. These results are broadly relevant to therapeutic gene editing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data are deposited at NCBI GEO accession GSE203560.
References
Field A, Adelman K. Evaluating enhancer function and transcription. Annu Rev Biochem. 2020;89:213–34.
de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24.
Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation center. Nature. 2012;485:381.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376.
Huang P, Keller CA, Giardine B, Grevet JD, Davies JOJ, Hughes JR, et al. Comparative analysis of three-dimensional chromosomal architecture identifies a novel fetal hemoglobin regulatory element. Genes Dev. 2017;31:1704–13.
Lettice LA, Daniels S, Sweeney E, Venkataraman S, Devenney PS, Gautier P, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32:1492–9.
Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428.
Agrawal P, Blinka S, Pulakanti K, Reimer MH, Stelloh C, Meyer AE, et al. Genome editing demonstrates that the −5 kb Nanog enhancer regulates Nanog expression by modulating RNAPII initiation and/or recruitment. J Biol Chem. 2021;296:100189.
Bolt CC, Lopez-Delisle L, Hintermann A, Mascrez B, Rauseo A, Andrey G, et al. Context-dependent enhancer function revealed by targeted inter-TAD relocation. bioRxiv. 2022;13:3488.
Kerschner JL, Paranjapye A, NandyMazumdar M, Yin S, Leir SH, Harris A. OTX2 regulates CFTR expression during endoderm differentiation and occupies 3’ cis-regulatory elements. Dev Dyn. 2021;250:684–700.
Gosalia N, Harris A. Chromatin dynamics in the regulation of CFTR expression. Genes (Basel). 2015;6:543.
Ensinck M, Mottais A, Detry C, Leal T, Carlon MS. On the corner of models and cure: gene editing in cystic fibrosis. Front Pharmacol. 2021;12:677.
Xia E, Duan R, Shi F, Seigel KE, Grasemann H, Hu J. Overcoming the undesirable CRISPR-Cas9 expression in gene correction. Mol Ther Nucleic Acids. 2018;13:699–709.
Zhou ZP, Yang LL, Cao H, Chen ZR, Zhang Y, Wen XY, et al. In vitro validation of a CRISPR-mediated CFTR correction strategy for preclinical translation in pigs. Hum Gene Ther. 2019;30:1101–16.
Vaidyanathan S, Baik R, Chen L, Bravo DT, Suarez CJ, Abazari SM, et al. Targeted replacement of full-length CFTR in human airway stem cells by CRISPR-Cas9 for pan-mutation correction in the endogenous locus. Mol Ther. 2022;30:223–37.
Suzuki S, Crane AM, Anirudhan V, Barillà C, Matthias N, Randell SH, et al. Highly efficient gene editing of cystic fibrosis patient-derived airway basal cells results in functional CFTR correction. Mol Ther. 2020;28:1684–95.
Bednarski C, Tomczak K, vom Hövel B, Weber WM, Cathomen T. Targeted integration of a super-exon into the CFTR locus leads to functional correction of a cystic fibrosis cell line model. PLoS One. 2016;11:e0161072.
Zhang Z, Leir SH, Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am J Respir Cell Mol Biol. 2015;52:387–96.
Zhang Z, Leir SH, Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol Cell Biol. 2013;33:2843–53.
Zhang Z, Ott CJ, Lewandowska MA, Leir SH, Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J Cell Mol Med. 2012;16:1321–30.
NandyMazumdar M, Yin S, Paranjapye A, Kerschner JL, Swahn H, Ge A, et al. Looping of upstream cis-regulatory elements is required for CFTR expression in human airway epithelial cells. Nucleic Acids Res. 2020;48:3513–24.
Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, et al. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;10:28–47.
Cai Z, Palmai-Pallag T, Khuituan P, Mutolo MJ, Boinot C, Liu B, et al. Impact of the F508del mutation on ovine CFTR, a Cl− channel with enhanced conductance and ATP-dependent gating. J Physiol. 2015;593:2427.
Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–62.
Joshi N, Fass JN. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33). 2011. https://github.com/najoshi/sickle.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60.
Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5.
Krijger PHL, Geeven G, Bianchi V, Hilvering CRE, de Laat W. 4C-seq from beginning to end: a detailed protocol for sample preparation and data analysis. Methods. 2020;170:17–32.
Fan Z, Perisse IV, Cotton CU, Regouski M, Meng Q, Domb C, et al. A sheep model of cystic fibrosis generated by CRISPR/Cas9 disruption of the CFTR gene. JCI Insight. 2018;3:e123529.
Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, et al. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci USA. 2009;106:19934–9.
Smith AN, Barth ML, McDowell TL, Moulin DS, Nuthall HN, Hollingsworth MA, et al. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J Biol Chem. 1996;271:9947–54.
Rowntree RK, Vassaux G, McDowell TL, Howe S, McGuigan A, Phylactides M, et al. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum Mol Genet. 2001;10:1455–64.
Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med. 2009;13:680–92.
Yin S, NandyMazumdar M, Paranjapye A, Harris A. Cross-talk between enhancers, structural elements and activating transcription factors maintains the 3D architecture and expression of the CFTR gene. Genomics. 2022;114:110350.
Lewandowska MA, Costa FF, Bischof JM, Williams SH, Soares MB, Harris A. Multiple mechanisms influence regulation of the cystic fibrosis transmembrane conductance regulator gene promoter. Am J Respir Cell Mol Biol. 2010;43:334–41.
Ramalho AS, Lewandowska MA, Farinha CM, Mendes F, Gonçalves J, Barreto C, et al. Deletion of CFTR translation start site reveals functional isoforms of the protein in CF patients. Cell Physiol Biochem. 2009;24:335–46.
Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A, et al. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016;44:3082–94.
Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem J. 2007;408:267–75.
Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–99.
Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genetics. 2016;48:176–82.
Valley HC, Bukis KM, Bell A, Cheng Y, Wong E, Jordan NJ, et al. Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J Cystic Fibrosis. 2019;18:476–83.
Acknowledgements
We thank Drs. Shih-Hsing Leir and Tom Kelley for helpful discussion and the Epithelial Cell Core at the CWRU CF Center (Cystic Fibrosis Foundation RDP R447-CR11) for Ussing chamber measurements. Also, Dr. Vian Peshdary for reagent generation, and the CWRU School of Medicine Genomics Core for sequencing.
Funding
This work was supported by the Cystic Fibrosis Foundation (Davis19XX0) and NIH HL094585 and HD068901 (both to AH).
Author information
Authors and Affiliations
Contributions
Conceptualization: JLK and AH. Methodology: JLK and AH. Validation: JLK and AP. Formal analysis: JLK and AP. Investigation: JLK, NV, and MDW. Data curation: AP. Writing – original draft: JLK and AH. Writing – review & editing: JLK and AH. Visualization: JLK. Supervision: AH. Project administration: AH. Funding acquisition: AH.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kerschner, J.L., Paranjapye, A., Vaghela, N. et al. An ectopic enhancer restores CFTR expression through de novo chromatin looping. Gene Ther 30, 478–486 (2023). https://doi.org/10.1038/s41434-022-00378-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-022-00378-7