Abstract
Anti-idiotype antibodies have been considered for vaccination approaches against different diseases, including cancers. Based on that, we previously described an anti-bevacizumab idiotype monoclonal antibody, 10.D7, that revealed detectable antitumor effects on a vascular endothelial growth factor (VEGF)-dependent tumor model. Herein, we evaluated the possible applicability of a single-chain variable fragment (scFv) for the 10.D7 antibody in a gene immunization strategy. After checking that mammalian cells transfected to express the 10.D7 scFv are recognized by bevacizumab, it was explored the ability of our scFv construction, in a gene-based scheme, to elicit an immune response containing VEGF-binding antibodies. The results provide evidence that the designed 10.D7 scFv construct maintains the anti-bevacizumab idiotype features and has potential to activate an immune response recognizing VEGF.
Similar content being viewed by others
Introduction
Based on the idiotype network theory [1], an anti-idiotype (Id) antibody reacts with the binding site of a particular antibody, presenting features that make it behave as an antigen surrogate. Anti-Id antibodies have been proposed to be used in vaccination against diseases, including those autoimmune [2], infectious [3], and neoplastic [4], especially for inducing antibody responses targeting proper or difficult-to-emulate conformational determinants [5,6,7]. Although several of the anti-Id applications use full-length monoclonal antibodies (mAbs) [8], polyclonal antibodies and immunoglobulin-derived recombinant formats have also been considered as immunogen [9]. An artificial antibody construction frequently used for that purpose is the single-chain variable fragment (scFv), which consists of the variable regions of the immunoglobulin heavy and light chains joined by a flexible linker [10].
The vascular endothelial growth factor (VEGF) is a self-glycoprotein known to participate in many physiologic and pathologic events [11]. This molecule is the main target of antiangiogenic strategies for cancer treatment. The first and most promising therapeutic product against a pro-angiogenic factor is a humanized anti-VEGF mAb, called bevacizumab. This antibody was initially approved by US FDA for use in colorectal carcinoma and, later, also in non-small cell lung carcinoma, cervical cancer, progressive glioblastoma, and renal cell carcinoma, among others [12].
We have formerly described an anti-bevacizumab idiotype mAb, 10.D7, that elicits VEGF-binding antibodies and shows considerable therapeutic potential in a tumor mouse model [13]. Herein, we report a proof-of-concept study aimed to evaluate whether an scFv for the 10.D7 mAb would maintain the anti-Id property of the corresponding full-length mAb and activate a VEGF-binding antibody response when used as an immunogen.
Materials and methods
Cell culture
HEK 293 T cells (ATCC, Manassas, VA, USA) were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA), and B16-F10 cells (ATCC) in RPMI-1640 (Thermo Fisher Scientific). Both media were supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific). 10.D7 hybridoma cells were maintained in RPMI-1640 containing 10% FBS and 50 μM 2-mercaptoethanol. All cells were cultured at 37 °C in a humidified atmosphere with 5% CO2 and were regularly checked for mycoplasma contamination.
Construction of 10.D7 scFv gene
Total RNA was isolated from 106 10.D7 hybridoma cells with TRIzol (Thermo Fisher Scientific). The cDNA of VH and VL were synthesized with oligos A and C, respectively, using High-capacity cDNA reverse transcription kit (Applied Biosystems, Waltham, MA, USA) (Table 1), as previously described [14]. Briefly, a solution containing 1 μg RNA in 5 μl of sterile water, 1 μl of 10 × buffer, 0.4 μl dNTP at 100 mM, 1 μl 10x random primer, 0.5 μl MultiScribe reverse transcriptase (Thermo Fisher Scientific) at 50 U/µl, and 2.1 µl of sterile water, totaling a 10-µl final volume, was prepared. The reaction was conducted in a thermocycler at 25 °C for 10 min, 37 °C for two hours, and 85 °C for 5 min.
The DNA fragments encoding for the VH and VL domains of 10.D7 mAb were then amplified by PCR using A + B and C + D oligo pairs (1 μM each), respectively (Table 1). PCR work solution was prepared according to the manufacturer’s instructions: 5 µl of cDNA diluted 1:100 in sterile water, 0.25 µl of sense primer and 0.25 µl of antisense, 12.5 µl of Master Mix (Thermo Fisher Scientific), and 7 µl of sterile water. The antibody heavy chain amplification conditions were: 94 °C for 3 min; 40 cycles at 94 °C for 1 min, 62 °C for 30 s, and 72 °C for 1 min; and a final incubation of 5 min at 72 °C. The light chain amplification conditions were: 94 °C for 3 min; 40 cycles at 94 °C for 1 min, 60 °C for 30 s and 72 °C for 1 min; 5 min at 72 °C. The products were analyzed by electrophoresis on a 1% agarose gel in TAE (40 mM Tris-acetate, 1 mM EDTA). The DNA bands were detected with SYBR Safe DNA stain (Thermo Fisher Scientific) on a Gel Doc EZ Imager system (Bio-Rad, Hercules, CA, USA). DNA fragments corresponding to VH and VL were purified from the agarose gel with Geneclean (MP Biomedicals, Solon, OH, USA) and further analyzed using BigDye Terminator Ready (Thermo Fisher Scientific) on a genetic analyzer sequencer. The 10.D7 scFv was designed in the VH-linker-VL orientation, using (Gly4Ser)3 as a linker [15], and was synthesized by a gene service (GenScript, Piscataway, NJ, USA) into a pcDNA3.1 expression vector containing the EcoRI and HindIII restriction sites. To confirm the final product, the plasmid vector was digested with EcoRI and HindIII, and the insert englobing the scFv gene was sequenced.
Animals
6–8-week-old male C57Bl/6 mice were obtained from “Centro de Desenvolvimento de Modelos Experimentais para Medicina e Biologia” (CEDEME/UNIFESP, Brazil) animal facility and maintained with a 12:12 h light:dark cycle and water ad libitum. Experiments were performed after anesthetic induction with ketamine (100 mg/kg; Syntec, Brazil) and xylazine (10 mg/kg; Syntec). Mice were euthanized by anesthetic overdose. All procedures were in accordance with the guidelines of the US National Research Council for care and use of laboratory animals [16].
Enzyme immunoassay (ELISA) studies
Cell-bound ELISA was performed to assess whether 10.D7 scFv gene-transfected cells are recognized by bevacizumab. For that, HEK 293 T cells were transfected with pcDNA3.1(+) containing or not the scFv gene, using the Superfect Transfection Reagent (Qiagen, Germany) following the manufacturer’s instructions. Transfected cells were plated on a 96-well plate (2 × 104 cells/well) and fixed with 0.05% glutaraldehyde (Sigma). Endogenous peroxidase was neutralized with 1% H2O2 (Sigma). After blocking with 1% bovine serum albumin (BSA; Sigma) in phosphate-buffered saline (PBS), wells were incubated for 1 h at 37 °C in the presence or absence of 1 µg/ml bevacizumab (Avastin; Roche, Switzerland). After washes with PBS containing 0.1% BSA and 0.05% Tween 20 (Sigma), wells were incubated with biotin-conjugated anti-human IgG (Sigma).
Indirect ELISA was carried out to detect VEGF-binding antibodies in serum samples. 96-well plates were coated with 50 ng/ml recombinant VEGF (Thermo Fisher Scientific), blocked with 1% BSA in PBS, and incubated overnight at 4 °C with bevacizumab (1 μg/ml) or serum samples from animals immunized with pcDNA3.1 or pcDNA3.1-scFv10.D7. Sera were obtained by bleeding the retro-orbital plexus of ketamine/xylazine-anesthetized mice 15 days after the last immunization dose. After washes, wells were incubated with anti-mouse or anti-human IgG secondary antibody (Thermo Fisher Scientific), both conjugated with biotin.
In both assays, after incubation with peroxidase-streptavidin (Sigma) for 30 min at room temperature, the wells were developed with o-phenylenediamine substrate (Sigma). The reaction was stopped with 4 N H2SO4 (Merck, Germany) and absorbance values were read at 490 nm.
Mouse immunization
For gene immunization studies, animals were intramuscularly injected with DNA plasmid (50 µg in each quadriceps) four times at 15-day intervals. Immediately after each administration, six electric pulses (100 V; 40 milliseconds per pulse; 1-second interval) were applied through 10-mm tweezer electrodes (T820-BTX; Genetronics, San Diego, CA, USA) positioned close to the injection sites [17, 18].
In vivo tumor growth
The potential antitumor effect of immunization with 10.D7 scFv gene was assessed in a subcutaneous tumor model. For that, on the 15th day after the last immunization dose, mice were subcutaneously challenged with 5 × 105 B16-F10 cells in the left flank. Tumor growth was monitored daily with a caliper. Two groups: pcDNA3.1-scFv10.D7-immunized, and pcDNA3.1-immunized (empty vector control) mice. Tumor volume was calculated considering the equation: Volume = (large diameter) × (small diameter)2 × 0.52 [19].
Statistical analyses
Statistical analyses were performed with GraphPad Prism, version 7.0 (GraphPad Software, La Jolla, CA, USA). Data were analyzed by Student’s t-test, when two groups were compared, or by one-way ANOVA followed by Bonferroni’s post-test, in the case of multiple comparisons, as indicated in the figure legends. The differences were considered significant when p < 0.05.
Results and discussion
Pre-clinical and clinical studies using plasmid DNA have been described to prevent or treat several diseases, including cancers, and their results are promising [20]. DNA is considered a valuable biomaterial to develop vaccines [21], with some attractive characteristics, such as its good stability. Also, DNA preparations are cost-effective and relative easy to be obtained compared to proteins [20].
In this work, we evaluated the potential of a DNA vaccine strategy, based on the idiotype network theory [1], to trigger immune response containing antibodies that bind to the vascular endothelial growth factor (VEGF). For that, the gene coding a single-chain variable fragment (scFv) [10] for the 10.D7 anti-idiotype (Id) mAb [13] was obtained. The genes of the variable (Fv) regions of the light (L) and heavy (H) chains were isolated from the 10.D7 hybridoma cells, sequenced, and used to construct a conventional scFv in the VH-VL orientation. Figure 1a shows 340-bp and 324-bp bands, referring to purified FvH and FvL products, respectively. BLAST analyses of these sequences showed ~94% and ~97% homology with other FvH and FvL mouse immunoglobulin chains, respectively.
a Gel electrophoresis of RT-PCR amplified products of VH and VL genes from 10.D7 hybridoma cells. VH- and VL-related bands have, respectively, ~340 and ~324 bp. A 1 kb Plus DNA ladder (Thermo Fisher Scientific) was used as a size marker. b Digestion products of pcDNA3.1-scFv10.D7 with EcoRI/HindIII (different clones). ~830-bp band insert, relative to a 100 bp DNA ladder (Promega, Madison, WI, USA), may comprise the 10.D7 scFv gene. Both analyses were performed on a 1% agarose gel.
The designed 10.D7 scFv gene was inserted along with a signal peptide into pcDNA3.1(+) vector to then functionally evaluate the encoded protein. As indicated in Fig. 1b, the digestion of the constructed plasmid DNA with EcoRI/Hindlll releases an insert of ~830-bp, which may correspond to the scFv gene. By using the obtained vector, our scFv gene was first checked whether it expresses a product recognized by bevacizumab. That analysis was performed by cell-bound ELISA in HEK 293 T cells transfected to express or not the 10.D7 scFv. Figure 2a shows that the bevacizumab binding to cells transfected with pcDNA3.1-scFv10.D7 plasmid is significantly higher than the detected in the empty vector group (p < 0.001; one-way ANOVA, followed by Bonferroni’s post-test). This result indicates that the designed scFv may preserve the bevacizumab-binding ability of the parental full-length mAb and, if any mismatched VH-VL arrangement occurred, which was already found for other scFvs [22, 23, 24], it does not seem to have affected the antibody anti-Id feature.
a Cell-bound ELISA detection of bevacizumab recognition of HEK 293 T cells transfected with pcDNA3.1-scFv10.D7. Cells transfected with pcDNA3.1 (empty vector) were used as control. Bevacizumab binding was detected with biotin-conjugated anti-human IgG antibodies followed by peroxidase-streptavidin. Assay performed in quadruplicate. Mean ± SD. **p < 0.001; one-way ANOVA/Bonferroni’s post-test. b In vivo experimental design. c ELISA detection of VEGF-binding antibodies in 1:100-diluted serum samples collected 15 days after the last immunization dose. Bevacizumab was used as a positive control. Negative control, pre-immune serum. Assay performed in quadruplicate. Mean ± SD. **p < 0.001; one-way ANOVA/Bonferroni’s post-test. d B16-F10 tumor growth curves. Mice were immunized with pcDNA3.1-scFv10.D7 (n = 9) or pcDNA3.1 (empty vector; n = 10). Tumor volume was calculated from the measured perpendicular diameters. Mean ± SD. *p < 0.005; **p < 0.001; Student’s t-test. e Tumor mass (mean ± SD) on the 14th day after tumor cell injection. Assay performed in quadruplicate. **p < 0.001; Student’s t-test.
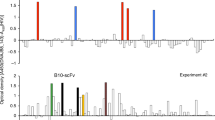
Thereafter, the designed 10.D7 scFv was explored as a DNA vaccine administered intramuscularly. In our approach, the scFv gene was delivered by electroporation, which was already reported to enhance the expression of plasmid genes [25] and elicit humoral immunity superior to that obtained when used the intramuscular route alone [26, 27]. It has been described the need for up to three doses to generate neutralizing antibodies sufficient to have detectable DNA vaccination efficacy [28]. We evaluated the presence of VEGF-binding antibodies after two and four doses, and the results, despite varying between animals, revealed in most cases the same antibody levels at both time points. However, we also had mouse whose antibody response was detected only in the last sample point considered (Supplementary Fig. 1), which led us to use in this work a four-dose protocol with the intention to maximize the number of responders. The vaccination protocol, provided in Fig. 2b, was designed based on the described previously [18].
VEGF-binding antibodies were assessed 15 days after the fourth immunization dose. For that, sera from C57BI/6 mice immunized with pcDNA3.1 or pcDNA3.1-scFv10.D7 vectors were analyzed by ELISA. The comparison between the experimental and empty vector control groups shows a significant difference (p < 0.001; one-way ANOVA/Bonferroni’s post-test) (Fig. 2c), which suggests that the expressed 10.D7 scFv may mimic VEGF and induces an immune response containing antibodies reactive to VEGF, which is a phylogenetically conserved molecule. Thus, this result indicates that the 10.D7 scFv immunization surpass an immune tolerance to a self-antigen [29].
The detected anti-anti-Id antibody levels were relatively low, which is not unexpected. The magnitude of the observed antibody response seems to be consistent with the one in the report used as a base for the 10.D7 scFv vaccination protocol. Like us, such work also showed modest ELISA results after the intramuscular-electroporation delivery of four doses of the gene of an anti-Id scFv that mimics a self-protein [18].
Despite the weak ELISA signals, it was enough to detect an antitumor activity. The application of the 10.D7 scFv gene in an immunization scheme was assessed in the B16-F10 subcutaneous tumor model, which is known to be dependent on VEGF [30]. Animals receiving the pcDNA3.1-scFv10.D7 plasmid showed reduced tumor growth compared to the empty vector control group (Fig. 2d). The average tumor mass at the end of the experiment, on the 14th day after tumor cell injection, was significantly lower in the scFv group than the detected in the control one (p = 0.0002; Student’s t-test) (Fig. 2e). These findings point out that the previously described antitumor effect resulting from full-length 10.D7 mAb immunization [13] is also detected following a 10.D7 scFv gene-based strategy, without the need to have a protein immunogen purified.
Our data bring evidence of a potential vaccine application of the gene of an anti-Id construction conceived from a commercial VEGF-targeting antibody, bevacizumab. Id vaccines have long been considered a treatment option for cancers [31]. The strategy not only has shown potential to influence tumor growth [32], but also has demonstrated some clinical benefits in phase I-III trials [33, 34], which reinforces the relevance of the Id approaches.
The gene vaccine development consists of a multistep process, going from the immunogen formulation format to the immunization regimen, which includes the gene delivery and the number of doses [21]. Although all these aspects can be optimized in the proposed anti-Id vaccine, the used protocol, based on a well-succeeded previous one [18], was useful to detect the potential of the 10.D7 scFv gene to trigger antibodies reacting with VEGF.
Together, our results reveal a promising scFv that may retain the anti-bevacizumab Id features of the previously described parental mAb. Based on the idiotype network, the designed construction has potential to be explored in DNA (or even mRNA) vaccines to activate VEGF-binding responses.
Data availability
The data that support the reported results are available from the corresponding authors upon reasonable request.
References
Jerne NK. Towards a network theory of the immune system. Ann Immunol (Paris). 1974;125C:373–89.
Pan SY, Chia YC, Yee HR, Fang Cheng AY, Anjum CE, Kenisi Y, et al. Immunomodulatory potential of anti-idiotypic antibodies for the treatment of autoimmune diseases. Future Sci OA. 2020;7:648. https://doi.org/10.2144/fsoa-2020-0142.
Cowton VM, Owsianka AM, Fadda V, Ortega-Prieto AM, Cole SJ, Potter JA, et al. Development of a structural epitope mimic: an idiotypic approach to HCV vaccine design. NPJ Vaccines. 2021;6:7. https://doi.org/10.1038/s41541-020-00269-1.
Stanova AK, Ryabkova VA, Utekhin SV, Shoenfeld VJ, Churilov LP, Shoenfeld Y. Anti-Idiotypic agonistic antibodies: candidates for the role of universal remedy. Antibodies (Basel). 2020;9:19. https://doi.org/10.3390/antib9020019.
Saha A, Chatterjee SK. Dendritic cells pulsed with an anti-idiotype antibody mimicking Her-2/neu induced protective antitumor immunity in two lines of Her-2/neu transgenic mice. Cell Immunol. 2010;263:9–21. https://doi.org/10.1016/j.cellimm.2010.02.010.
Ladjemi MZ, Chardes T, Corgnac S, Garambois V, Morisseau S, Robert B, et al. Vaccination with human anti-trastuzumab anti-idiotype scFv reverses HER2 immunological tolerance and induces tumor immunity in MMTV.f.huHER2(Fo5) mice. Breast Cancer Res. 2011;13:17. https://doi.org/10.1186/bcr2826.
Kohler H, Pashov A, Kieber-Emmons T. The promise of anti-idiotype revisited. Front Immunol. 2019;10:808. https://doi.org/10.3389/fimmu.2019.00808.
Ladjemi MZ. Anti-idiotypic antibodies as cancer vaccines: achievements and future improvements. Front Oncol. 2012;2:158. https://doi.org/10.3389/fonc.2012.00158.
Ascoli CA, Aggeler B. Overlooked benefits of using polyclonal antibodies. Biotechniques. 2018; https://doi.org/10.2144/btn-2018-0065.
Bemani P, Mohammadi M, Hakakian A. ScFv improvement approaches. Protein Pept Lett. 2018; https://doi.org/10.2174/0929866525666171129225436.
Ye X, Gaucher JF, Vidal M, Broussy S. A structural overview of vascular endothelial growth factors pharmacological ligands: from macromolecules to designed peptidomimetics. Molecules. 2021; https://doi.org/10.3390/molecules26226759.
Garcia J, Hurwitz HI, Sandler AB, Miles D, Coleman RL, Deurloo R, et al. Bevacizumab (Avastin®) in cancer treatment: a review of 15 years of clinical experience and future outlook. Cancer Treat Rev. 2020;86:102017. https://doi.org/10.1016/j.ctrv.2020.102017.
Sanches Jde S, de Aguiar RB, Parise CB, Suzuki JM, Chammas R, de Moraes JZ. Anti-bevacizumab idiotype antibody vaccination is effective in inducing vascular endothelial growth factor-binding response, impairing tumor outgrowth. Cancer Sci. 2016; https://doi.org/10.1111/cas.12903.
Pignatari GC, Takeshita D, Parise CB, Soares FA, de Moraes JZ, Han SW. Carcinoembryonic antigen (CEA) mimicry by an anti-idiotypic scFv isolated from anti-Id 6.C4 hybridoma. J Biotechnol. 2007;127:615–25. https://doi.org/10.1016/j.jbiotec.2006.08.007.
de Aguiar RB, da Silva TA, Costa BA, Machado MFM, Yamada RY, Braggion C, et al. Generation and functional characterization of a single-chain variable fragment (scFv) of the anti-FGF2 3F12E7 monoclonal antibody. Sci Rep. 2021; https://doi.org/10.1038/s41598-020-80746-8.
National Research Council, Guide for the Care and Use of Laboratory Animals, 8th ed., Nat Acad Press,Washington (DC), 2011.
Li L, Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev Vaccines. 2016; https://doi.org/10.1586/14760584.2016.1124762.
Denapoli PMA, Zanetti BF, Dos Santos AA, de Moraes JZ, Han SW. Preventive DNA vaccination against CEA-expressing tumors with anti-idiotypic scFv6.C4 DNA in CEA-expressing transgenic mice. Cancer Immunol Immunother. 2017;66:333–42. https://doi.org/10.1007/s00262-016-1940-4.
de Aguiar RB, Parise CB, Souza CR, Braggion C, Quintilio W, Moro AM, et al. Blocking FGF2 with a new specific monoclonal antibody impairs angiogenesis and experimental metastatic melanoma, suggesting a potential role in adjuvant settings. Cancer Lett. 2016;371:151–60. https://doi.org/10.1016/j.canlet.2015.11.030.
Liu MA. DNA vaccines: an historical perspective and view to the future. Immunol Rev. 2011;239:62–84. https://doi.org/10.1111/j.1600-065X.2010.00980.x.
Jorritsma SHT, Gowans EJ, Grubor-Bauk B, Wijesundara DK. Delivery methods to increase cellular uptake and immunogenicity of DNA vaccines. Vaccine. 2016;34:5488–94. https://doi.org/10.1016/j.vaccine.2016.09.062.
Francisco JA, Gilliland LK, Stebbins MR, Norris NA, Ledbetter JA, Siegall CB. Activity of a single-chain immunotoxin that selectively kills lymphoma and other B-lineage cells expressing the CD40 antigen. Cancer Res. 1995;55:3099–104.
Tripathi PK, Qin H, Deng S, Xu C, Bhattacharya-Chatterjee M, Foon KA, et al. Antigen mimicry by an anti-idiotypic antibody single chain variable fragment. Mol Immunol. 1998;35:853–63. https://doi.org/10.1016/s0161-5890(98)00072-8.
Liu A, Ye Y, Chen W, Wang X, Chen F. Expression of V(H)-linker-V(L) orientation-dependent single-chain Fv antibody fragment derived from hybridoma 2E6 against aflatoxin B1 in Escherichia coli. J Ind Microbiol Biotechnol. 2015; https://doi.org/10.1007/s10295-014-1570-9.
Mir LM, Bureau MF, Gehl J, Rangara R, Rouy D, Caillaud JM, et al. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc Natl Acad Sci USA. 1999;96:4262–7. https://doi.org/10.1073/pnas.96.8.4262.
Parise CB, Lisboa B, Takeshita D, Sacramento CB, de Moraes JZ, Han SW. Humoral immune response after genetic immunization is consistently improved by electroporation. Vaccine. 2008;26:3812–7. https://doi.org/10.1016/j.vaccine.2008.05.029.
Lee YH, Lim H, Lee JA, Kim SH, Hwang YH, In HJ, et al. Optimization of Zika DNA vaccine by delivery systems. Virology. 2021;559:10–14. https://doi.org/10.1016/j.virol.2021.03.005.
Fomsgaard A, Liu MA. The key role of nucleic acid vaccines for one health. Viruses. 2021;13:258. https://doi.org/10.3390/v13020258.
Coelho M, Gauthier P, Pugnière M, Roquet F, Pèlegrin A, Navarro-Teulon I. Isolation and characterisation of a human anti-idiotypic scFv used as a surrogate tumour antigen to elicit an anti-HER-2/neu humoral response in mice. Br J Cancer. 2004;90:2032–41. https://doi.org/10.1038/sj.bjc.6601825.
Ghosh S, Maity P. Augmented antitumor effects of combination therapy with VEGF antibody and cisplatin on murine B16F10 melanoma cells. Int Immunopharmacol. 2007;7:1598–608. https://doi.org/10.1016/j.intimp.2007.08.017.
Grimmett E, Al-Share B, Alkassab MB, Zhou RW, Desai A, Rahim MMA, et al. Cancer vaccines: past, present and future; a review article. Discov Oncol. 2022;13:31 https://doi.org/10.1007/s12672-022-00491-4.
Inogès S, Rodrìguez-Calvillo M, Zabalegui N, Lòpez-Dìaz de Cerio A, Villanueva H, Soria E, et al. Clinical benefit associated with idiotypic vaccination in patients with follicular lymphoma. J Natl Cancer Inst. 2006;98:1292–301. https://doi.org/10.1093/jnci/djj358.
Schuster SJ, Neelapu SS, Gause BL, Janik JE, Muggia FM, Gockerman JP, et al. Vaccination with patient-specific tumor-derived antigen in first remission improves disease-free survival in follicular lymphoma. J Clin Oncol. 2011;29:2787–94. https://doi.org/10.1200/JCO.2010.33.3005.
Meleshko AN, Petrovskaya NA, Savelyeva N, Vashkevich KP, Doronina SN, Sachivko NV. Phase I clinical trial of idiotypic DNA vaccine administered as a complex with polyethylenimine to patients with B-cell lymphoma. Hum Vaccin Immunother. 2017;13:1–6. https://doi.org/10.1080/21645515.2017.1285477.
Acknowledgements
This work was supported by São Paulo Research Foundation (FAPESP; Grant Number 16/14358-2), and the Brazilian National Research Council (CNPq).
Author information
Authors and Affiliations
Contributions
RBA and JZM conceived the experiments. TAS and GEM performed the assays. BH offered technical support. MM and MFMM contributed to scFv construction. All authors analyzed the results. RBA and JZM wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Animal procedures were approved by the ethics committee (CEUA) of the “Universidade Federal de São Paulo” (Protocol no. 6710130416).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Silva, T.A., Aguiar, R.B., Mori, M. et al. Potential of an anti-bevacizumab idiotype scFv DNA-based immunization to elicit VEGF-binding antibody response. Gene Ther 30, 598–602 (2023). https://doi.org/10.1038/s41434-022-00376-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-022-00376-9