Abstract
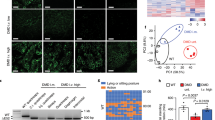
With the development of basic research, some genetic-based methods have been found to treat Duchenne muscular dystrophy (DMD) with large deletion mutations and nonsense mutations. Appropriate therapeutic approaches for repairing multiple duplications are limited. We used the CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 system with patient-derived primary myoblasts to correct multiple duplications of the dystrophin gene. Muscle tissues from a patient carrying duplications of dystrophin were obtained, and tissue-derived primary cells were cultured. Myoblasts were purified with an immunomagnetic sorting system using CD56 microbeads. After transduction by lentivirus with a designed single guide RNA (sgRNA) targeting a duplicated region, myoblasts were allowed to differentiate for 7 days. Copy number variations in the exons of the patient’s myotubes were quantified by real-time PCR before and after genetic editing. Western blot analysis was performed to detect the full-length dystrophin protein before and after genetic editing. The ten sequences predicted to be the most likely off-targets were determined by Sanger sequencing. The patient carried duplications of exon 18–25, dystrophin protein expression was completely abrogated. Real-time PCR showed that the copy number of exon 25 in the patient’s myotubes was 2.015 ± 0.079 compared with that of the healthy controls. After editing, the copy number of exon 25 in the patient’s modified myotubes was 1.308 ± 0.083 compared with that of the healthy controls (P < 0.001). Western blot analysis revealed no expression of the dystrophin protein in the patient’s myotubes before editing. After editing, the patient’s myotubes expressed the full-length dystrophin protein at a level that was ~6.12% of that in the healthy control samples. Off-target analysis revealed no abnormal editing at the ten sites predicted to be the most likely off-target sites. The excision of multiple duplications by the CRISPR/Cas9 system restored the expression of full-length dystrophin. This study provides proof of evidence for future genome-editing therapy in patients with DMD caused by multiple duplication mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data generated or analyzed during this study are presented in the main paper and the supplementary file. Additional data are available from the corresponding author on reasonable request.
References
Falzarano MS, Scotton C, Passarelli C, Ferlini A. Duchenne muscular dystrophy: from diagnosis to therapy. Molecules. 2015;20:18168–84.
Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, et al. The treat-nmd dmd global database: analysis of more than 7,000 duchenne muscular dystrophy mutations. Hum Mutat. 2015;36:395–402.
Matthews E, Brassington R, Kuntzer T, Jichi F, Manzur AY. Corticosteroids for the treatment of duchenne muscular dystrophy. Cochrane Database Syst Rev. 2016:CD003725.
Hsu PD, Lander ES, Zhang F. Development and applications of crispr-cas9 for genome engineering. Cell. 2014;157:1262–78.
Goell JH, Hilton IB. Crispr/cas-based epigenome editing: Advances, applications, and clinical utility. Trends Biotechnol. 2021;39:678–91.
Vazquez ME, Cabarcos MR, Roman TD, Stein AJ, Garcia ND, Nazar BA, et al. Cellular cardiomyoplasty: Development of a technique to culture human myoblasts for clinical transplantation. Cell Tissue Bank. 2005;6:117–24.
Tassin A, Leroy B, Laoudj-Chenivesse D, Wauters A, Vanderplanck C, Le Bihan MC, et al. Fshd myotubes with different phenotypes exhibit distinct proteomes. PLoS ONE. 2012;7:e51865.
Schumm M, Lang P, Bethge W, Faul C, Feuchtinger T, Pfeiffer M, et al. Depletion of t-cell receptor alpha/beta and cd19 positive cells from apheresis products with the clinimacs device. Cytotherapy. 2013;15:1253–8.
Suttmann H, Jacobsen M, Reiss K, Jocham D, Bohle A, Brandau S. Mechanisms of bacillus calmette-guerin mediated natural killer cell activation. J Urol. 2004;172:1490–5.
Lattanzi A, Duguez S, Moiani A, Izmiryan A, Barbon E, Martin S, et al. Correction of the exon 2 duplication in dmd myoblasts by a single crispr/cas9 system. Mol Ther Nucleic Acids. 2017;7:11–9.
Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex crispr/cas9-based genome editing for correction of dystrophin mutations that cause duchenne muscular dystrophy. Nat Commun. 2015;6:6244.
Xu L, Park KH, Zhao L, Xu J, El Refaey M, Gao Y, et al. Crispr-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 2016;24:564–9.
Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–3.
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, et al. In vivo genome editing improves muscle function in a mouse model of duchenne muscular dystrophy. Science. 2016;351:403–7.
Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–11.
Kole R, Krieg AM. Exon skipping therapy for duchenne muscular dystrophy. Adv Drug Deliv Rev. 2015;87:104–7.
Greer KL, Lochmuller H, Flanigan K, Fletcher S, Wilton SD. Targeted exon skipping to correct exon duplications in the dystrophin gene. Mol Ther Nucleic Acids. 2014;3:e155.
Li X, Zhao L, Zhou S, Hu C, Shi Y, Shi W, et al. A comprehensive database of duchenne and becker muscular dystrophy patients (0–18 years old) in east China. Orphanet J Rare Dis. 2015;10:5.
Tong YR, Geng C, Guan YZ, Zhao YH, Ren HT, Yao FX, et al. A comprehensive analysis of 2013 dystrophinopathies in china: A report from national rare disease center. Front Neurol. 2020;11:572006.
Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, et al. Large deletions induced by cas9 cleavage. Nature. 2018;560:E8–E9.
Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, et al. Crispr-cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun. 2019;10:1136.
Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, Sauna ZE. Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol Ther Methods Clin Dev. 2018;10:105–12.
Hakim CH, Kumar SRP, Perez-Lopez DO, Wasala NB, Zhang D, Yue Y, et al. Cas9-specific immune responses compromise local and systemic AAV CRISPR therapy in multiple dystrophic canine models. Nat Commun. 2021;12:6769.
Gee P, Lung MSY, Okuzaki Y, Sasakawa N, Iguchi T, Makita Y, et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat Commun. 2020;11:1334.
Funding
This work was supported by the grant 81801248 from the National Natural Science Foundation of China, grant 81870902 from the National Natural Science Foundation of China, and the Joint Funds 2018Y9082 for the Innovation of Science and Technology of Fujian province.
Author information
Authors and Affiliations
Contributions
NW conceived and initiated the research; NW and Z-QW directed the research; NW and D-NW designed the experiments; D-NW and M-TL generated and characterized reagents and tools; D-NW performed all the experiments; D-NW and Z-QW analyzed the data together with all authors; D-NW wrote the paper with the aid of NW. All the authors read and approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, DN., Wang, ZQ., Jin, M. et al. CRISPR/Cas9-based genome editing for the modification of multiple duplications that cause Duchenne muscular dystrophy. Gene Ther 29, 730–737 (2022). https://doi.org/10.1038/s41434-022-00336-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-022-00336-3