Abstract
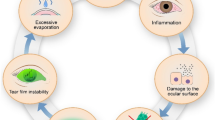
Corneal and ocular surface diseases (OSDs) carry significant psychosocial and economic burden worldwide. We set out to review the literature on the application of artificial intelligence (AI) and bioinformatics for analysis of biofluid biomarkers in corneal and OSDs and evaluate their utility in clinical decision making. MEDLINE, EMBASE, Cochrane and Web of Science were systematically queried for articles using AI or bioinformatics methodology in corneal and OSDs and examining biofluids from inception to August 2021. In total, 10,264 articles were screened, and 23 articles consisting of 1058 individuals were included. Using various AI/bioinformatics tools, changes in certain tear film cytokines that are proinflammatory such as increased expression of apolipoprotein, haptoglobin, annexin 1, S100A8, S100A9, Glutathione S-transferase, and decreased expression of supportive tear film components such as lipocalin-1, prolactin inducible protein, lysozyme C, lactotransferrin, cystatin S, and mammaglobin-b, proline rich protein, were found to be correlated with pathogenesis and/or treatment outcomes of dry eye, keratoconus, meibomian gland dysfunction, and Sjögren’s. Overall, most AI/bioinformatics tools were used to classify biofluids into diseases subgroups, distinguish between OSD, identify risk factors, or make predictions about treatment response, and/or prognosis. To conclude, AI models such as artificial neural networks, hierarchical clustering, random forest, etc., in conjunction with proteomic or metabolomic profiling using bioinformatics tools such as Gene Ontology or Kyoto Encylopedia of Genes and Genomes pathway analysis, were found to inform biomarker discovery, distinguish between OSDs, help define subgroups with OSDs and make predictions about treatment response in a clinical setting.
摘要
角膜和眼表疾病 (OSDs) 在世界范围内带来了严重的心理和经济负担。我们对人工智能 (AI) 和生物信息学分析角膜和眼表疾病中生物流体标志物的相关文献进行了综述, 评估其在临床决策中的实用性。 本文对MEDLINE、EMBASE、Cochrane和Web of Science数据库进行系统筛选, 找到从建库到2021年8月期间将人工智能或生物信息学方法应用于角膜和眼表疾病并检查了生物体液的文章。共筛选10264篇文献, 纳入23篇, 共1058位受试者。 使用各种人工智能/生物信息学工具发现某些促炎性泪膜细胞因子的表达变化, 例如载脂蛋白、结合珠蛋白、膜联蛋白1、S100A8、S100A9和谷胱甘肽S-转移酶的表达增加, 支持性泪膜成分如脂质运载蛋白-1、催乳素诱导蛋白、溶菌酶C、乳转铁蛋白、胱抑素S的表达减少, 乳球蛋白-B和脯氨酸富集类蛋白等与干眼症、圆锥角膜、睑板腺功能障碍和干燥综合征的发病机制和/或治疗结果相关。总之, 大部分人工智能/生物信息学工具可根据生物液体将疾病亚群进行分类, 从而区分眼表疾病, 识别风险因素或对治疗反应和/或预后进行预测。 总的来说, 人工智能模型如人工神经网络, 分层聚类, 随机森林等, 在进行蛋白质组或代谢组分析中使用的生物信息学工具, 如基因本体论或京都基因和基因组路径分析百科全书, 可以为生物标志物的发现提供信息, 区分眼表疾病, 帮助定义眼表疾病亚群, 并在临床环境中预测治疗反应。
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 18 print issues and online access
$259.00 per year
only $14.39 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
von Thun und Hohenstein-Blaul N, Funke S, Grus FH. Tears as a source of biomarkers for ocular and systemic diseases. Exp Eye Res. 2013;117:126–37.
Zhou L, Beuerman RW. Tear analysis in ocular surface diseases. Progr Retinal Eye Res. 2012;31:527–50.
Wang MTM, Muntz A, Wolffsohn JS, Craig JP. Association between dry eye disease, self-perceived health status, and self-reported psychological stress burden. Clin Exp Optom. 2021;104:835–40.
Yu J, Asche CV, Fairchild CJ. The economic burden of dry eye disease in the United States: A decision tree analysis. Cornea. 2011;30:379–87.
Chan C, Ziai S, Myageri V, Burns JG, Prokopich CL. Economic burden and loss of quality of life from dry eye disease in Canada. BMJ Open Ophthalmol. 2021;6:e000709.
Yang W, Luo Y, Wu S, Niu X, Yan Y, Qiao C, et al. Estimated annual economic burden of dry eye disease based on a multi-center analysis in china: a retrospective study. Front Med (Lausanne). 2021;8:771352.
de Almeida Borges D, Alborghetti MR, Franco Paes Leme A, Ramos Domingues R, Duarte B, Veiga M, et al. Tear proteomic profile in three distinct ocular surface diseases: keratoconus, pterygium, and dry eye related to graft-versus-host disease. Clin Proteomics [Internet]. 2020;17:1–16.
Ji YW, Kim HM, Ryu SY, Oh JW, Yeo A, Choi CY, et al. Changes in human tear proteome following topical treatment of dry eye disease: Cyclosporine a versus diquafosol tetrasodium. Invest Ophthalmol Vis Sci. 2019;60:5035–44.
Menegay M, Lee DM, Tabbara KF, Cafaro TA, Urrets-Zavalía JA, Serra HM, et al. Proteomic analysis of climatic keratopathy droplets. Invest Ophthalmol Vis Sci. 2008;49:2829–37.
Jiang Y, Yang C, Zheng Y, Liu Y, Chen Y. A set of global metabolomic biomarker candidates to predict the risk of dry eye disease. Front Cell Dev Biol. 2020;8:344.
Wojakowska A, Pietrowska M, Widlak P, Dobrowolski D, Wylegała E, Tarnawska D. Metabolomic signature discriminates normal human cornea from Keratoconus—A pilot GC/MS study. Molecules. 2020;25:2933.
González N, Iloro I, Soria J, Duran JA, Santamaría A, Elortza F, et al. Human tear peptide/protein profiling study of ocular surface diseases by SPE-MALDI-TOF mass spectrometry analyses. EuPA Open Proteom. 2014;3:206–15.
Soria J, Villarrubia A, Merayo-Lloves J, Elortza F, Azkargorta M, de Toledo JA, et al. Label-free LC–MS/MS quantitative analysis of aqueous humor from keratoconic and normal eyes. Mol Vis. 2015;21:451–60.
Keskinbora K, Güven F. Artificial intelligence and ophthalmology. Turk J Ophthalmol. 2020;50:37–43.
Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunović H. Artificial intelligence in retina. Prog Retin Eye Res. 2018;67:1–29.
Yu LR, Stewart NA, Veenstra TD. Chapter 8 - Proteomics: The Deciphering of the Functional Genome. In: Ginsburg GS, Willard HFBTE of G and PM, editors. San Diego: Academic Press; 2010:89–96
Larrañaga P, Calvo B, Santana R, Bielza C, Galdiano J, Inza I, et al. Machine learning in bioinformatics. Brief Bioinform. 2006;7:86–112.
Cryan LM, O’Brien C. Proteomics as a research tool in clinical and experimental ophthalmology. Proteomics Clin Appl. 2008;2:762–75.
Schmidt A, Forne I, Imhof A. Bioinformatic analysis of proteomics data. BMC Syst Biol. 2014;8(Suppl 2):S3–S3. 2014/03/13
Tan SZ, Begley P, Mullard G, Hollywood KA, Bishop PN. Introduction to metabolomics and its applications in ophthalmology. Eye (Basingstoke). 2016;30:773–83.
Hopkins JJ, Keane PA, Balaskas K. Delivering personalized medicine in retinal care: From artificial intelligence algorithms to clinical application. Curr Opin Ophthalmol. 2020;31:329–36.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71.
Munn Z, Moola S, Riitano D, Lisy K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int J Health Policy Manag. 2014;3:123–8.
Valesan LF, Da-Cas CD, Réus JC, Denardin ACS, Garanhani RR, Bonotto D, et al. Prevalence of temporomandibular joint disorders: a systematic review and meta-analysis. Clin Oral Investig. 2021;25:441–53.
Liang JT, Huang J, Chen TC, Hung JS. The Toldt fascia: A historic review and surgical implications in complete mesocolic excision for colon cancer. Asian J Surg. 2019;42:1–5.
Srinivasan S, Thangavelu M, Zhang L, Green KB, Nichols KK. iTRAQ quantitative proteomics in the analysis of tears in dry eye patients. Invest Ophthalmol Vis Sci. 2012;53:5052–9.
Huang Z, Du CX, Pan XD. The use of in-strip digestion for fast proteomic analysis on tear fluid from dry eye patients. PLoS One. 2018;13:e0200702.
Yawata N, Awate S, Liu YC, Yuan S, Woon K, Siak J, et al. Kinetics of tear fluid proteins after endothelial keratoplasty and predictive factors for recovery from corneal haze. J Clin Med. 2020;9:1–14.
Linghu D, Guo L, Zhao Y, Liu Z, Zhao M, Huang L, et al. iTRAQ-based quantitative proteomic analysis and bioinformatics study of proteins in pterygia. 2018;1600094:7–8.
Aqrawi LA, Galtung HK, Vestad B, Øvstebø R, Thiede B, Rusthen S, et al. Identification of potential saliva and tear biomarkers in primary Sjögren’s syndrome, utilising the extraction of extracellular vesicles and proteomics analysis. Arthritis Res Ther. 2017;19:1–15.
Grus FH, Podust VN, Bruns K, Lackner K, Fu S, Dalmasso EA, et al. SELDI-TOF-MS ProteinChip array profiling of tears from patients with dry eye. Invest Ophthalmol Vis Sci. 2005;46:863–76.
Tong L, Zhou L, Beuerman R, Simonyi S, Hollander DA, Stern ME. Effects of punctal occlusion on global tear proteins in patients with dry eye. Ocular Surface. 2017;15:736–41.
Zou X, Wang S, Zhang P, Lu L, Zou H. Quantitative proteomics and weighted correlation network analysis of tear samples in adults and children with diabetes and dry eye. Transl Vis Sci Technol. 2020;9:1–15.
Soria J, Durán JA, Etxebarria J, Merayo J, González N, Reigada R, et al. Tear proteome and protein network analyses reveal a novel pentamarker panel for tear film characterization in dry eye and meibomian gland dysfunction. J Proteomics. 2013;78:94–112.
Fodor M, Vitályos G, Losonczy G, Hassan Z, Pásztor D, Gogolák P, et al. Tear mediators NGF along with IL-13 predict keratoconus progression. Ocul Immunol Inflamm. 2021;29:1090–101.
Fodor M, Gogolák P, Rajnavölgyi É, Berta A, Kardos L, Módis L, et al. Long-term kinetics of cytokine responses in human tears after penetrating keratoplasty. J Interferon Cytokine Res. 2009;29:375–9.
Kim SW, Lee J, Lee B, Rhim T. Proteomic analysis in pterygium; upregulated protein expression of ALDH3A1, PDIA3, and PRDX2. Mol Vis. 2014;20:1192–202.
Sembler-Møller ML, Belstrøm D, Locht H, Pedersen AML. Proteomics of saliva, plasma, and salivary gland tissue in Sjögren’s syndrome and non-Sjögren patients identify novel biomarker candidates. J Proteomics. 2020;225:103877.
O’leary OE, Schoetzau A, Amruthalingam L, Geber-Hollbach N, Plattner K, Jenoe P, et al. Tear proteomic predictive biomarker model for ocular graft versus host disease classification. Transl Vis Sci Technol. 2020;9:1–15.
Piyacomn Y, Kasetsuwan N, Reinprayoon U, Satitpitakul V, Tesapirat L. Efficacy and safety of intense pulsed light in patients with meibomian gland dysfunction-a randomized, double-masked, sham-controlled clinical trial. Cornea. 2020;39:325–32.
Leonardi A, Palmigiano A, Mazzola EA, Messina A, Milazzo EMS, Bortolotti M, et al. Identification of human tear fluid biomarkers in vernal keratoconjunctivitis using iTRAQ quantitative proteomics. Allergy: Eur J Allergy Clin Immunol. 2014;69:254–60.
Sembler-Møller ML, Belstrøm D, Locht H, Pedersen AML. Proteomics of saliva, plasma, and salivary gland tissue in Sjögren’s syndrome and non-Sjögren patients identify novel biomarker candidates. J Proteomics. 2020;225:103877.
Inamoto Y, Valdés-Sanz N, Ogawa Y, Alves M, Berchicci L, Galvin J, et al. Ocular graft-versus-host disease after hematopoietic cell transplantation: Expert review from the Late Effects and Quality of Life Working Committee of the CIBMTR and Transplant Complications Working Party of the EBMT. Bone Marrow Transpl. 2019;54:662–73.
Yam GHF, Fuest M, Zhou L, Liu YC, Deng L, Chan ASY, et al. Differential epithelial and stromal protein profiles in cone and non-cone regions of keratoconus corneas. Sci Rep. 2019;9:1–17.
Sharif R, Bak-Nielsen S, Sejersen H, Ding K, Hjortdal J, Karamichos D. Prolactin-induced protein is a novel biomarker for keratoconus. Exp Eye Res. 2019;179:55–63.
Soria J, Acera A, Merayo-Lloves J, Durán JA, González N, Rodriguez S, et al. Tear proteome analysis in ocular surface diseases using label-free LC-MS/MS and multiplexed-microarray biomarker validation. Sci Rep. 2017;7:17478.
Karimpour-Fard A, Elaine Epperson L, Hunter LE. A survey of computational tools for downstream analysis of proteomic and other omic datasets. Hum Genomics. 2015;9:28.
Lancashire LJ, Lemetre C, Ball GR. An introduction to artificial neural networks in bioinformatics—Application to complex microarray and mass spectrometry datasets in cancer studies. Briefings in Bioinformatics. 2009;10:315–29.
González N, Iloro I, Durán JA, Elortza F, Suárez T. Evaluation of inter-day and inter-individual variability of tear peptide/protein profiles by MALDI-TOF MS analyses. Mol Vis. 2012;18:1572–82.
Vizcaíno JA, Foster JM, Martens L. Proteomics data repositories: Providing a safe haven for your data and acting as a springboard for further research. J Proteomics. 2010;73:2136.
Khatri P, Drǎghici S. Ontological analysis of gene expression data: Current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–95.
Green-Church KB, Nichols KK, Kleinholz NM, Zhang L, Nichols JJ. Investigation of the human tear film proteome using multiple proteomic approaches. Mol Vis. 2008;14:456–70.
Grus FH, Sabuncuo P, Augustin AJ. Analysis of tear protein patterns of dry-eye patients using fluorescent staining dyes and two-dimensional quantification algorithms. Electrophoresis. 2001;22:1845–50.
Edorh NA, el Maftouhi A, Djerada Z, Arndt C, Denoyer A. New model to better diagnose dry eye disease integrating OCT corneal epithelial mapping. Br J Ophthalmol. 2021;106:1488–95.
Acknowledgements
This study was in-part funded by a Fighting Blindness Canada research grant awarded to Dr. Tina Felfeli. The authors would like to acknowledge Arshpreet Bassi, Shaily Brahmbhatt, Priyanka Singh, Ishita Aggarwal, Amy Basilious, Jasmine Bhatti, and Karthik Manickavachagam, who participated in article screening.
Author information
Authors and Affiliations
Contributions
TF, RNM: conceptualization, supervision, methodology, writing, review and editing of manuscript. DRP: conceptualization, data collection, analysis and interpretation of results, draft manuscript preparation. SHK, methodology, data collection. AP, data collection, analysis and interpretation of the results. All authors reviewed the results, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Search strategy used to survey databases.
Appendix B
Assessment of risk of bias and quality of included studies based on criteria from Joanna Briggs Institute Critical Appraisal Tools.
Supplementary information
41433_2022_2307_MOESM2_ESM.docx
Assessment of risk of bias and quality of included studies based on criteria from Joanna Briggs Institute Critical Appraisal Tools
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pur, D.R., Krance, S.H., Pucchio, A. et al. Current uses of artificial intelligence in the analysis of biofluid markers involved in corneal and ocular surface diseases: a systematic review. Eye 37, 2007–2019 (2023). https://doi.org/10.1038/s41433-022-02307-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41433-022-02307-9
This article is cited by
-
Is tear proteome profile a predictor of developing uveitis in ANA-positive patients with oligoarticular juvenile idiopathic arthritis?
Graefe's Archive for Clinical and Experimental Ophthalmology (2024)
-
Applications of artificial intelligence and bioinformatics methodologies in the analysis of ocular biofluid markers: a scoping review
Graefe's Archive for Clinical and Experimental Ophthalmology (2023)