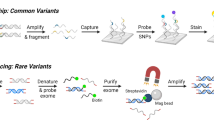
Abstract
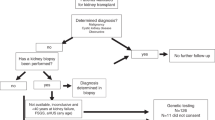
KiT-GENIE is a monocentric DNA biobank set up to consolidate the very rich and homogeneous DIVAT French cohort of kidney donors and recipients (D/R) in order to explore the molecular factors involved in kidney transplantation outcomes. We collected DNA samples for kidney transplantations performed in Nantes, and we leveraged GWAS genotyping data for securing high-quality genetic data with deep SNP and HLA annotations through imputations and for inferring D/R genetic ancestry. Overall, the biobank included 4217 individuals (n = 1945 D + 2,272 R, including 1969 D/R pairs), 7.4 M SNPs and over 200 clinical variables. KiT-GENIE represents an accurate snapshot of kidney transplantation clinical practice in Nantes between 2002 and 2018, with an enrichment in living kidney donors (17%) and recipients with focal segmental glomerulosclerosis (4%). Recipients were predominantly male (63%), of European ancestry (93%), with a mean age of 51yo and 86% experienced their first graft over the study period. D/R pairs were 93% from European ancestry, and 95% pairs exhibited at least one HLA allelic mismatch. The mean follow-up time was 6.7 years with a hindsight up to 25 years. Recipients experienced biopsy-proven rejection and graft loss for 16.6% and 21.3%, respectively. KiT-GENIE constitutes one of the largest kidney transplantation genetic cohorts worldwide to date. It includes homogeneous high-quality clinical and genetic data for donors and recipients, hence offering a unique opportunity to investigate immunogenetic and genetic factors, as well as donor-recipient interactions and mismatches involved in rejection, graft survival, primary disease recurrence and other comorbidities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The KiT-GENIE data are available from our CR2TI team. Data and summary statistics are available upon reasonable request following approval from the KiT-GENIE steering committee and ethics committee to ensure data protection and privacy in compliance with French and European laws. Collaborations are encouraged through specific research projects using the KiT-GENIE data or through enriching the existing cohort with new patients. Potential collaborators are invited to contact the primary investigator Sophie Limou: sophie.limou@univ-nantes.fr.
References
Hill NR, Fatoba ST, Oke JL, Hirst JA, O’Callaghan CA, Lasserson DS, et al. Global prevalence of chronic kidney disease - a systematic review and meta-analysis. PLoS ONE. 2016;11:e0158765.
Dalrymple LS, Katz R, Kestenbaum B, Shlipak MG, Sarnak MJ, Stehman-Breen C, et al. Chronic kidney disease and the risk of end-stage renal disease versus death. J Gen Intern Med. 2011;26:379–85.
Garcia GG, Harden P, Chapman J. The global role of kidney transplantation. Kidney Blood Press Res. 2012;35:299–304.
Tonelli M, Wiebe N, Knoll G, Bello A, Browne S, Jadhav D, et al. Systematic review: kidney transplantation compared with dialysis in clinically relevant outcomes. Am J Transpl. 2011;11:2093–109.
Wong G, Howard K, Chapman JR, Chadban S, Cross N, Tong A, et al. Comparative survival and economic benefits of deceased donor kidney transplantation and dialysis in people with varying ages and co-morbidities. PLoS ONE. 2012;7:e29591.
Agence de la Biomédecine. Annual report 2020—Kidney transplantation. 2021. https://rams-archives2020.agence-biomedecine.fr/sites/default/files/pdf/2021-08/ABM_PG_Organes_Rein2020.pdf.
Gondos A, Döhler B, Brenner H, Opelz G. Kidney graft survival in Europe and the United States. Transpl J 2013;95:267–74.
Sasaki N, Idica A The HLA-matching effect in different cohorts of kidney transplant recipients: 10 years later. Clin Transpl. 2010;261‑82.
Tam V, Patel N. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–84.
Levy SE, Myers RM. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet. 2016;17:95–115.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9.
Hanif Z, Sufiyan N, Patel M, Akhtar MZ. Role of biobanks in transplantation. Ann Med Surg. 2018;28:30–3.
Cambon-Thomsen A, Ducournau P, Gourraud PA, Pontille D. Biobanks for Genomics and Genomics for Biobanks. Comp Funct Genomics. 2003;4:628–34.
Alaa AM, Bolton T, Di Angelantonio E, Rudd JHF, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: A prospective study of 423,604 UK Biobank participants. PLOS ONE. 2019;14:e0213653. Aalto-Setala K, Aalto-Setala K, éditeurs.
Xu X, Eales JM, Jiang X, Sanderson E, Drzal M, Saluja S, et al. Contributions of obesity to kidney health and disease: insights from Mendelian randomization and the human kidney transcriptomics. Cardiovascular Res. 2022;118:3151–61.
Yu Z, Jin J, Tin A, Köttgen A, Yu B, Chen J, et al. Polygenic risk scores for kidney function and their associations with circulating proteome, and incident kidney diseases. J Am Soc Nephrol. 2021;32:3161–73.
Ghisdal L, Baron C, Lebranchu Y, Viklický O, Konarikova A, Naesens M, et al. Genome-wide association study of acute renal graft rejection. Am J Transpl. 2017;17:201–9.
Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, Mollon J, Delaney F, Perucha E, et al. Long- and short-term outcomes in renal allografts with deceased donors: a large recipient and donor genome-wide association study. Am J Transpl. 2018;18:1370–9.
Massart A, Ghisdal L, Viklicky O, Naesens M, Abramowicz D, Abramowicz M. Reply to Hernandez et al.—GWAS of acute renal graft rejection. Am J Transplant. 2018;18:2098–9.
Pihlstrøm HK, Mjøen G, Mucha S, Haraldsen G, Franke A, Jardine A, et al. Single nucleotide polymorphisms and long-term clinical outcome in renal transplant patients: a validation study. Am J Transpl. 2017;17:528–33.
Reindl-Schwaighofer R, Heinzel A, Signorini L, Thaunat O, Oberbauer R. Mechanisms underlying human genetic diversity: consequence for antigraft antibody responses. Transpl Int. 2018;31:239–50.
Steers NJ, Li Y, Drace Z, D’Addario JA, Fischman C, Liu L, et al. Genomic mismatch at LIMS1 locus and kidney allograft rejection. N Engl J Med. 2019;380:1918–28.
Reindl-Schwaighofer R, Heinzel A, Kainz A, van Setten J, Jelencsics K, Hu K, et al. Contribution of non-HLA incompatibility between donor and recipient to kidney allograft survival: genome-wide analysis in a prospective cohort. Lancet. 2019;393:910–7.
Ba R, Geffard E, Douillard V, Simon F, Mesnard L, Vince N, et al. Surfing the big data wave: omics data challenges in transplantation. Transplantation. 2022;106:e114–25.
Zanoni F, Kiryluk K. Genetic background and transplantation outcomes: insights from genome-wide association studies. Curr Opin Organ Transpl. 2020;25:35–41.
InternationalGenetics & Translational Research in Transplantation Network (iGeneTRAiN).Design and implementation of the international genetics and translational research in transplantation network.Transplantation.2015;99:2401–12.
Masset C, Kerleau C, Garandeau C, Ville S, Cantarovich D, Hourmant M, et al. A third injection of the BNT162b2 mRNA COVID-19 vaccine in kidney transplant recipients improves the humoral immune response. Kidney Int. 2021;100:1132–5.
Foucher Y, Lorent M, Albano L, Roux S, Pernin V, Le Quintrec M, et al. Renal transplantation outcomes in obese patients: a French cohort-based study. BMC Nephrol. 2021;22:79.
Dujardin A, Chesneau M, Dubois F, Danger R, Bui L, Kerleau C, et al. Clinical and immunological follow-up of very long-term kidney transplant recipients treated with calcineurin inhibitors indicates dual phenotypes. Kidney Int. 2021;99:1418–29.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75.
Browning BL, Tian X, Zhou Y, Browning SR. Fast two-stage phasing of large-scale sequence data. Am J Hum Genet. 2021;108:1880–90.
Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14:192–200.
Byrska-Bishop M, Evani US, Zhao X, Basile AO, Abel HJ, Regier AA, et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell. 2022;185:3426–40.e19.
Douillard V, Castelli EC, Mack SJ, Hollenbach JA, Gourraud PA, Vince N, et al. Approaching genetics through the MHC Lens: tools and methods for HLA research. Front Genet. 2021;12:774916.
Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39.
Olson E, Geng J, Raghavan M. Polymorphisms of HLA-B: influences on assembly and immunity. Curr Opin Immunol. 2020;64:137–45.
Mesnard L, Muthukumar T, Burbach M, Li C, Shang H, Dadhania D, et al. Exome sequencing and prediction of long-term kidney allograft function. PLoS Comput Biol. 2016;12:e1005088.
O’Brien RP, Phelan PJ, Conroy J, O’Kelly P, Green A, Keogan M, et al. A genome-wide association study of recipient genotype and medium-term kidney allograft function. Clin Transpl. 2013;27:379–87.
Divers J, Ma L, Brown WM, Palmer ND, Choi Y, Israni AK, et al. Genome‐wide association study for time to failure of kidney transplants from African American deceased donors. Clin Transpl. 2020;34:e13827.
Lam NN, Lloyd A, Lentine KL, Quinn RR, Ravani P, Hemmelgarn BR, et al. Changes in kidney function follow living donor nephrectomy. Kidney Int. 2020;98:176–86.
Mjøen G, Hallan S, Hartmann A, Foss A, Midtvedt K, Øyen O, et al. Long-term risks for kidney donors. Kidney Int 2014;86:162–7.
Muzaale AD, Massie AB, Wang MC, Montgomery RA, McBride MA, Wainright JL, et al. Risk of end-stage renal disease following live kidney donation. JAMA. 2014;311:579–86.
Rudnicki M. FSGS recurrence in adults after renal transplantation. BioMed Res Int. 2016;2016:1–7.
Hanks SC, Forer L, Schönherr S, LeFaive J, Martins T, Welch R, et al. Extent to which array genotyping and imputation with large reference panels approximate deep whole-genome sequencing. Am J Hum Genet. 2022;109:1653–66.
Fishman CE, Mohebnasab M, van Setten J, Zanoni F, Wang C, Deaglio S, et al. Genome-wide study updates in the international genetics and translational research in transplantation network (iGeneTRAiN). Front Genet. 2019;10:1084.
Acknowledgements
We thank everyone who helped in the design, collection, genotyping experiments, data cleaning and analyses. We are especially indebted to all donors and recipients who participated in this study, to the clinical staff and physicians who care and manage the patients on a daily basis and helped recruiting the patients, and to research associates who participated in the data collection. Clinical data were collected from the French DIVAT multicentric prospective cohort of kidney and/or pancreatic transplant recipients by focusing on the Nantes centre (www.divat.fr, n° CNIL 914184, ClinicalTrials.gov identifier: NCT02900040). The analysis and interpretation of these data are the responsibility of the authors. We would like to acknowledge the local blood bank and HLA typing lab (Etablissement Français du Sang [EFS] Nantes) for facilitating the access to DNA samples. Finally, we thank the GenoBiRD and Curie genomic platforms for technical support and GWAS genotyping.
Funding
The KiT-GENIE cohort has been funded by several entities: (1) The Etoiles Montantes funding by the Pays de la Loire region (n°2018-09998), (2) The IRCT Dialyse research project by the Société Francophone de Néphrologie, Dialyse et Transplantation (SFNDT), (3) The Greffe research project by French Agence de la Biomédecine (ABM, n°18GREFFE014). In addition, this work has benefited from government support through the National Research Agency (ANR) under the future investment program (n°ANR-17-RHUS-0010) and from the European Union’s Horizon 2020 Research and Innovation Programme (n°754995). Finally, we thank the Roche Pharma, Novartis, Astellas, Chiesi, Sandoz and Sanofi laboratories for supporting the DIVAT cohort as the CENTAURE Foundation (http://www.fondation-centaure.org).
Author information
Authors and Affiliations
Contributions
RB, AD and SL were involved in the design, conception, implementation and data collection of KiT-GENIE. RB, AD, CC, SLBB, SS, PG, and CP were involved in DNA collection and normalisation. RB, AD, VM, MM, VD, OR and SL were involved in quality controls, statistical description, imputation of missing data, PCA analysis and ancestry analysis. CK realized clinical data extraction from the DIVAT database. MG and GB provided clinical guidance and ensured access to the DIVAT clinical data. RB and SL wrote the article. SL, PAG and NV supervised the building of KiT-GENIE, revised and formalized the scientific content of the article and mentored RB. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Written consent was obtained from each participant for access to clinical data and participation to DNA analyses (DIVAT n° CNIL 914184). Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ba, R., Durand, A., Mauduit, V. et al. KiT-GENIE, the French genetic biobank of kidney transplantation. Eur J Hum Genet 31, 1291–1299 (2023). https://doi.org/10.1038/s41431-023-01294-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01294-z
This article is cited by
-
Deep phenotyping and population-level data can help resolve genomic variants
European Journal of Human Genetics (2023)