Abstract
Heterozygous single nucleotide variants (SNVs) or copy-number variant deletions involving FOXF1 or its distant lung-specific enhancer on chromosome 16q24.1 have been identified in 80–90% of patients with Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV), a lethal neonatal lung developmental disorder. We describe a four-generation family with a deceased ACDMPV neonate, her sibling from the electively terminated pregnancy, healthy mother with a history of pulmonary arterial hypertension (PAH), an unaffected aunt, an aunt deceased due to findings consistent with ACDMPV, and a reportedly unaffected grandmother, all with the frameshifting variant c.881_902dup (p.Gly302Profs*46) in FOXF1, and a deceased great-grandmother with a history of PAH. Genome sequencing analyses in the proband’s unaffected mother revealed a non-coding putative regulatory SNV rs560517434-A within the lung-specific distant FOXF1 enhancer in trans to the FOXF1 frameshift mutation. Functional testing of this variant using an in vitro luciferase reporter assay showed that it increased FOXF1 promoter activity 10-fold. Our studies further demonstrate that non-coding SNVs in the FOXF1 enhancer region can rescue the lethal ACDMPV phenotype and support the compound inheritance gene dosage model.
Similar content being viewed by others
Introduction
Lethal lung developmental disorders (LLDDs) are rare diseases of neonates manifesting with severe progressive respiratory failure and persistent pulmonary arterial hypertension (PAH). Based on histopathological appearance at lung biopsy or autopsy, they have been classified into Alveolar capillary dysplasia with misalignment of the pulmonary veins (ACDMPV; MIM# 265380), Acinar dysplasia (AcDys), Congenital alveolar dysplasia (CAD), and other unspecified primary pulmonary hypoplasias (PHs) [1, 2].
The majority of patients with AcDys or CAD have been found to have alterations involving TBX4, FGF10, or FGFR2, whereas greater than 80–90% of histopathologically-verified ACDMPV patients have heterozygous single nucleotide variants (SNVs) or copy-number variant (CNV) deletions involving the transcription factor (TF) Forkhead Box 1 gene (FOXF1; MIM# 601089) and/or its lung-specific enhancer located ~ 270 kb upstream to the gene on chromosome 16q24.1 [3,4,5].
In LLDD infants, we have identified a statistically significant enrichment of non-coding SNVs, within the lung-specific enhancers, located in trans to pathogenic heterozygous variants involving FOXF1, TBX4, or FGF10 that might act as hyper- or hypomorphs dramatically changing the critical gene expression levels and thus, modifying the LLDD and PAH phenotype, implying a complex compound inheritance gene dosage (CIGD) model of LLDDs [6,7,8,9,10].
Here, we describe clinical, histopathological, and molecular findings in a four-generation family with an ACDMPV with a segregating FOXF1 frameshifting variant. We propose that a non-coding hypermorphic SNV located in trans within the affected FOXF1 allele might have rescued the lethal ACDMPV phenotype in the proband’s mother and aunt.
Methods
Human subjects
Blood samples from the deceased neonate proband (IV:1), her parents (III:1 and III:2), grandparents (II:1 and II:2), and the unaffected aunt (III:4), and the formalin-fixed paraffin-embedded lung tissue obtained at autopsy from the proband (IV:1) and her deceased aunt (III:3) were collected after obtaining written informed consent (Table 1, Fig. 1 and Supplementary Fig. 1).
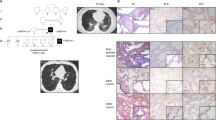
A The pathogenic segregating frameshifting FOXF1 variant c.881_902dup (p.Gly302Profs*46) is depicted as an asterix. The hypermorphic non-coding SNV rs560517434 in the distant lung-specific enhancer region upstream to FOXF1 is shown as A. Proband (IV:1) and her aunt (III:3) (black shade) died due to ACDMPV at the age two and three days, respectively; the proband’s mother (III:2) and maternal great-grandmother (I:1) (vertical lines) had pulmonary arterial hypertension at birth. The other family members have no abnormal lung phenotype. B Reporter assay showing about 10-fold increase of FOXF1 promoter activity by the enhancer SNV rs560517434-A (One-way ANOVA with post-hoc Tuckey HSD test, compared to the sole promoter or promoter with the common variant G, p ≤ 0.01). Data represent means of triplicate experiments ± SD. Data points are shown with dots.
Next generation sequencing
DNA extraction, clinical exome sequencing (ES) and genome sequencing (GS) are detailed in Supplementary Information.
Luciferase reporter assay
The functional significance of the candidate regulatory non-coding SNV, rs560517434-A, was assessed by sub-cloning it next to the FOXF1 promoter previously inserted into a luc2 reporter plasmid pGL4.10 (Promega, Madison, WI) and using this construct in the luciferase reporter assay in fetal lung fibroblasts IMR-90 (ATCC, Manassas, VA,) as described in [6] and Supplementary Information. Statistical significance was calculated by One-way ANOVA with post-hoc Tuckey HSD test.
Results
Clinical findings
The proband (IV:1) was born by an emergency Caesarean section at 28 + 6 weeks gestation following a pregnancy complicated by grade 4 placenta praevia and antepartum haemorrhages at 26 and 28 weeks. The mother received a complete course of antenatal steroids at first presentation, with a rescue dose 2 days prior to delivery. APGAR scores were 6, 4, and 9. She was placed on CPAP initially, was intubated at 2 h of age for increasing oxygen requirements and received 3 doses of surfactant. She deteriorated at 24 h of age and was clinically diagnosed as persistent pulmonary hypertension of the newborn. She developed systemic hypotension and jaundice of prematurity, and despite escalation of respiratory support she passed away on day 4.
Her mother (III:2) was born at 31 weeks gestation and developed pulmonary hypertension and bilateral pneumothoraces. These were treated successfully, and she has not had any significant medical issues since. A recent echocardiogram was unremarkable. A maternal aunt (III:3) was born at term following an unremarkable pregnancy. From 14 h of age, she experienced recurrent episodes of cyanosis and hypoxia which could not be managed with ventilation and she passed away at 3 days of age. Her maternal great-grandmother is reported to have had PAH as an adult.
Histopathological studies
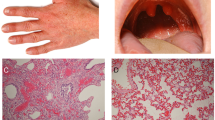
A review of the lung histopathology from the proband (IV:1) fetopsy showed diffuse bilateral lymphangiectasia with thickened alveolar septa and pulmonary veins juxtaposed with pulmonary arterioles and distal bronchioles. The pulmonary arteries were slightly thickened and accompanied by dilated congested veins. There were a few veins within interlobular septa. The “alveolar” walls had marked decreased vasculature. The CD31 showed enhanced septal capillarization. The D2-40 was weak but showed a mild increase in lymphatics within the pleura and possibly accounting for some of the cystic spaces within the parenchyma (Supplementary Fig. 2A, B).
Re-evaluation of the slides from lung autopsy performed in the proband’s deceased aunt (III:3) revealed thickened alveolar septa lined by plump pneumocytes, centrally placed capillaries and misaligned pulmonary venules. The lungs were very immature, arrested in the canalicular to saccular stage of development with airspaces lined by cuboidal epithelium (Supplementary Fig. 2C, D).
Molecular and computational analyses
Family trio ES revealed in the proband (IV:1) a heterozygous frameshift variant c.881_902dup (p.Gly302Profs*46; NM_001451.3) in exon 1 of FOXF1, inherited from the mother (III:2). Using Sanger sequencing, this variant was also found in the proband’s grandmother (II:1), sibling (IV:2, electively terminated pregnancy), maternal deceased aunt (III:3), and unaffected aunt (III:4) (Table 1, Fig. 1 and Supplementary Fig. 1). This variant is absent in the gnomAD (v2.1.1) and (v3.1.2) databases and predicted to truncate FOXF1 by replacing 78 amino acid C-terminal sequence (~24% of protein) with the new 45 amino acids.
GS analyses in the proband’s unaffected mother revealed a non-coding SNV rs560517434-A (gnomAD minor allele frequency (MAF): 0.1%), in the ~ 7 kb interval (chr16:86,252,422-86,258,902; hg19) within the ~ 60 kb distant FOXF1 lung-specific enhancer region mapping upstream to FOXF1 [4, 6] in the vicinity of the previously reported other putative hypermorphic variants mitigating ACDMPV lethality [6]. This variant is also present in her healthy sister and absent in the proband and 22 ACDMPV patients with the 16q24.1 CNV deletions (Table 1, Fig. 1 and Supplementary Fig. 1). This SNV is located within the binding site of the RAD21 and EZH2 TFs (ENCODE). The reporter assay in lung fibroblast cells IMR-90 showed that, in comparison to the common variant G of rs560517434, the A allele increased the FOXF1 promoter activity ~10-fold (p ≤ 0.01) (Fig. 1).
GS analysis of the ~60 kb lung-specific enhancer region in the grandmother (II:1) revealed 31 non-coding SNVs with variant allele fraction >0.2 that are absent in the proband. Thirteen of them are also absent in the 12 ACDMPV patients with 16q24.1 deletion (Supplementary Table 1). Two out of 31 detected non-coding SNVs, rs79338875-T (MAF: 7%) and rs74618630-T (MAF: 7%) are located in the ~ 7 kb core enhancer interval (Supplementary Fig. 1). However, reporter assay showed no significant difference in the FOXF1 promoter activity (Supplementary Fig. 3).
Discussion
The pLI score 0.96 for FOXF1 indicates that it is almost intolerant to loss-of-function variants. Thus, the observed variable expressivity and incomplete penetrance of ACDMPV suggests the involvement of the modifiers mitigating disease manifestation in the proband’s mother, aunt, maternal grandmother, and potentially great-grandmother. Based on our previous results and the CIGD model for LLDDs [6,7,8,9,10], we have considered a possibility of a modifier in trans to FOXF1 in the proband’s mother and the unaffected aunt inherited from their father and absent in the affected aunt, and a different modifier in the grandmother. Alternatively, in the grandmother (and likely in the great-grandmother) other modifier(s) on the different chromosome(s) might have been transmitted to her two unaffected daughters and not to the deceased daughter. However, too small pedigree precludes obtaining an informative LOD score in the latter scenario.
Recently Szafranski et al. reported three rare hypermorphic non-coding SNVs in trans to the heterozygous pathogenic SNV and CNV deletions involving FOXF1 and/or its lung-specific enhancer that likely mitigated the lethal ACDMPV phenotype [6]. Consistent with the complex compound inheritance reported by Karolak et al. [7], most recently, Yıldız Bölükbaşı et al. described a non-coding SNV located in trans to frameshift variant in TBX4 within the putative lung-specific enhancer in a multi-generation family with a wide spectrum of phenotypes, acting as a hypomorphic modifier [9].
Here, we describe the rare non-coding SNVs rs560517434-A, located within the ~7 kb core enhancer interval of the distant FOXF1 lung-specific enhancer in trans to the FOXF1-truncating variant c.881_902dup, and propose that it acts as a hypermorph, substantially alleviating the phenotypes in the proband’s mother and aunt. In an in vitro assay, it increased the activity of the FOXF1 promoter 10-fold, suggesting that it might have increased expression of FOXF1 on the wild-type allele in trans (Fig. 2). This variant is located within the ChIP-seq-determined EZH2 and RAD21 binding sites and might have modified stringency of their binding to DNA. Interestingly, this variant is in close proximity to the previously reported other putative hypermorphic variants mitigating ACDMPV lethality [6] (Fig. 2).
The identified rare (MAF = 0.1%) hypermorphic non-coding SNV rs560517434-A mapping to the ~ 7 kb interval (chr16:86,252,422-86,258,902; hg19) is located in trans to the pathogenic c.881_902dup (p.Gly302Profs*46; NM_001451.3) variant in exon 1 of FOXF1 on the other allele. Luciferase reporter assay revealed that rs560517434-A increased the FOXF1 promoter activity ~10-fold when compared to the common wildtype variant G. The previously reported hypermorphic non-coding SNVs [6] are shown in light gray. One of them, rs150502618-A, increased the FOXF1 promoter activity in the luciferase reporter assay 2.5-fold.
However, in the proband’s unaffected grandmother, we have not identified any candidate variant in the ~60 kb lung-specific enhancer region that might have rescued the ACDMPV phenotype. Thirteen SNVs present in the proband’s grandmother and absent in the proband and 12 ACDMPV patients with 16q24.1 CNV deletion involving the ~60 kb lung-specific enhancer region, are not located in the conserved region and most of them do not overlap any TF binding site although some of them may have regulatory function and warrant further investigation. Thus, it is more likely that a putative modifier maps to a different genomic region.
Unfortunately, due to an insufficient amount of poor-quality DNA sample we were not able to unambiguously conclude whether the younger ACDMPV sibling of the mother (III:3) had rs560517434-A.
Nevertheless, although episomal reporter assay is a powerful tool to access the effect of an enhancer region on the promoter by measuring luciferase gene expression, it also has some limitations. It assumes a priori that a tested interval is in vivo in immediate vicinity of the promoter. Moreover, it can only test a limited region of an enhancer or a variant that has been taken out of native chromatin context.
In summary, our studies provide further functional evidence that rare non-coding SNVs present within the distant regulatory region of FOXF1 might mitigate the expressivity of lethal ACDMPV phenotype and confirm the CIGD model of involvement of the coding and the non-coding variants in LLDDs.
Data availability
This variant has been submitted to the ClinVar database under the submission number SUB10968355. The data that support this study are available from the corresponding author upon reasonable request.
References
Nogee LM. Interstitial lung disease in newborns. Semin Fetal Neonatal Med. 2017;22:227–33.
Vincent M, Karolak JA, Deutsch G, Gambin T, Popek E, Isidor B, et al. Clinical, histopathological, and molecular diagnostics in lethal lung developmental disorders. Am J Respir Crit Care Med. 2019;200:1093–101.
Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–91.
Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O, et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23–33.
Szafranski P, Gambin T, Dharmadhikari AV, Akdemir KC, Jhangiani SN, Schuette J, et al. Pathogenetics of alveolar capillary dysplasia with misalignment of pulmonary veins. Hum Genet. 2016;135:569–86.
Szafranski P, Liu Q, Karolak JA, Song X, de Leeuw N, Faas B, et al. Association of rare non-coding SNVs in the lung-specific FOXF1 enhancer with a mitigation of the lethal ACDMPV phenotype. Hum Genet. 2019;138:1301–11.
Karolak JA, Vincent M, Deutsch G, Gambin T, Cogné B, Pichon O, et al. Complex compound inheritance of lethal lung developmental disorders due to disruption of the TBX-FGF pathway. Am J Hum Genet. 2019;104:213–28.
Karolak JA, Gambin T, Honey EM, Slavik T, Popek E, Stankiewicz P. A de novo 2.2 Mb recurrent 17q23.1q23.2 deletion unmasks novel putative regulatory non-coding SNVs associated with lethal lung hypoplasia and pulmonary hypertension: a case report. BMC Med Genomics. 2020;13:34.
Yıldız Bölükbaşı E, Karolak JA, Szafranski P, Gambin T, Murik O, Zeevi DA, et al. Exacerbation of mild lung disorders to lethal pulmonary hypoplasia by a noncoding hypomorphic SNV in a lung-specific enhancer in trans to the frameshifting TBX4 variant. Am J Med Genet A 2022;188:1420–5.
Lupski JR. Biology in balance: human diploid genome integrity, gene dosage, and genomic medicine. Trends Genet. 2022;38:554–71.
Acknowledgements
We would like to thank Dr. E. Popek for the histopathological evaluations and the family for participating and supporting this study.
Funding
Design of the study and collection, analysis, and interpretation of data were supported by grant awarded by the US National Institutes of Health (NIH), National Heart Lung and Blood Institute (NHLBI) R01HL137203 (PS). The Genomic Autopsy Study research was supported by NHMRC grant (APP1123341), Genomics Health Futures Mission—Medical Research Futures Fund (GHFM76777), and the Australian Genomic Health Alliance NHMRC Targeted Call for Research into Preparing Australia for the Genomics Revolution in Healthcare (GNT1113531) (HS and CB). PA was supported by fellowships from The Hospital Research Foundation and the Royal Adelaide Hospital Research Foundation.
Author information
Authors and Affiliations
Contributions
EYB executed the experiments; EYB, PSz, JAK, TG, PS analyzed and interpreted the data; AM did the histopathalogical evaluations; SM, HSS, PA, TH, CPB and JR provided clinical material; SM, AM, HSS, PA, TH, CPB and JR interpreted and described clinical findings; EYB, JAK, PS wrote the manuscript; all authors reviewed and discussed the manuscript during preparation and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The initial diagnosis in the proband was performed as part of the NHMRC and GHFM-MRFF funded Genomic Autopsy Study and was approved by the Human Ethics Committee of the Women’s and Children’s Health Network, South Australia, Australia (HREC/15/WCHN/35) and the Melbourne Health Human Research Ethics Committee as part of the Australian Genomics Health Alliance protocol: HREC/16/MH/251. Informed consent for genomic analysis and participation in study protocols was obtained from parents, and all research was conducted in accordance with the Declaration of Helsinki. The study research protocols were approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine (H-8712). Informed consent was obtained from all participants prior to genetic testing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yıldız Bölükbaşı, E., Karolak, J.A., Szafranski, P. et al. Variable expressivity in a four-generation ACDMPV family with a non-coding hypermorphic SNV in trans to the frameshifting FOXF1 variant. Eur J Hum Genet 30, 1182–1186 (2022). https://doi.org/10.1038/s41431-022-01159-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-022-01159-x
This article is cited by
-
Happy 30th birthday to the European Journal of Human Genetics!
European Journal of Human Genetics (2022)