Abstract
CTCF germline mutations have been related to MRD21. We report the first bilateral Wilms tumor suffered by a MRD21 patient carrying an unreported CTCF missense variant in a zinc finger domain of CTCF protein. We found that germline heterozygous variant I446K became homozygous in the tumor due to a loss of heterozygosity rearrangement affecting the whole q arm on chromosome 16. Our findings propose CTCF I446K variant as a link between MRD21 and Wilms tumor predisposition.
Similar content being viewed by others
Introduction
Several molecular alterations are recurrently detected in Wilms tumors (WT), but 11p aberrations have a prominent role. WT1 and several genes placed in 11p15.5 locus are commonly dysfunctional in Nephroblastoma [1, 2]. IGF2 is located at this locus and, it is commonly overexpressed in sporadic WT [2]. The Imprinting Control Center (ICR) is a 2.4 Kb in length region positioned between IGF2 and H19 gene (H19-ICR) which controls the expression of both genes. Physiologically, maternal IGF2 allele is silenced through a refined imprinting regulation carried out on this region. ICR DNA is unmethylated on the maternal allele and methylated on the paternally derived allele. IGF2 can be expressed when ICR is methylated and therefore, it occurs only in the paternal allele. Conversely, H19 is only expressed from the unmethylated maternal allele. This accurate regulation requires the presence of wild type CTCF protein [3]. This pattern of regulation on 11p15.5 locus is usually disrupted in WT. The somatic biallelic expression of IGF2 in WT can be induced as a result of two independent mechanisms: (1) Duplication of the paternal allele by LOH (loss of heterozygosity) and paternal uniparental disomy or (2) Increased ICR methylation on maternal allele (Loss of imprinting; LOI) [4].
A broad spectrum of constitutional genetic mutations, genomic aberrations and epigenetic deregulation are known to predispose to WT development. The Knudson’s two-hit hypothesis fits correctly for different genetic syndromes which include WT in their phenotype [1]. Among them highlight those associated with WT1 mutations or deletions [5] and Beckwith-Wiedemann syndrome (BWS).
Constitutional CTCF mutations, including missense mutations within zinc-finger domains, have been detected among patients with Mental Retardation Autosomal Dominant 21 (MRD21; OMIM #615502) [6]. These patients usually exhibit a short stature, microcephaly, intellectual disability with a broad clinical spectrum, minor facial dysmorphism and cardiac anomalies [7,8,9]. Complete CTCF loss of function in germline is lethal in mice during embryonal development and probably in humans as well [10]. Heterozygous germline variants in CTCF gene have not been previously related to WT risk.
Materials and methods
NGS-based gene panel was Pediatric-OncoPanelDx (by Imegen). DNA was isolated from blood with RecoverAll kit (Invitrogen) and from frozen tumor-selected section with QIAamp DNA Investigator Kit (QIAGEN). DNA quantification and integrity were assessed with Qubit and TapeStation. Libraries were prepared with Agilent SureSelect customized panel (254 genes, 0.8 Mb, Agilent XT-HS). Sequencing by Illumina NextSeq at 2 x 150 bp; Depth > 1000X. Bioinformatic analysis was performed with BWA-MEM aligner, caller variant VarDict and annotation by Ensembl Variant Effect Predictor. Quality metrics performed through Picard Pipeline (Broad Institute).
RNA was isolated from the paraffined tumor using RNeasy Mini Kit (Qiagen) and the retrotranscription with Taqman reverse transcription reagents (Applied Biosystems) and primers 5´TGTGCGATTACGCCAGTGTAGA3´; 5´GGCTCCTCCTCATCCTCATTGT3´.
DNA isolated from frozen tumor was analyzed by molecular karyotyping with SNPa (Cytoscan HD, Affymetrix). SNPa results were analyzed with Chromosome Analysis Suite software (Affymetrix, ChAS; version 3.1; GRCh37 (hg19)). SNPa data quality was assessed with the internal array quality control parameter “Median of the Absolute Values of all Pairwise Differences”. SNPa data were plotted and interpreted as previously described [11].
We selected 4 microsatellites in 16q22.1 around CTCF locus (D16S3107, D16S3085, D16S421, D16S3086) and amplified them by PCR using fluorescently labeled primers that flank the repeated sequence. The labeled PCR products were subsequently analyzed by capillary electrophoresis (Abi Prism 3130XL) to separate the amplicons by size.
Results
Patient data
Male patient was born on September 14th 2016 to Caucasian parents. The father previously suffered a seminoma with lung metastasis. Mother´s pregnancy was normal without any detectable ultrasound alterations. The child was born with an age-appropriate weight, height and cranial perimeter. Bilateral cryptorchidism was observed and, a bilateral renal ectasia (grade II) confirmed the first days of life. During neonatal age he was admitted in hospital due to pathological hypoglycemia. A heart murmur was auscultated and, an interatrial communication type oval fossa diagnosed. He also presented a mild ascending aortic ectasia. During the next months he grew normally with psychomotor development at the lower limit of normal range. He also presented a constitutionally minor facial dysmorphia consisting on a prominent forehead, bushy and arched eyebrows, long philtrum and thin upper lip (Fig. 1A).
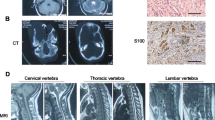
A Phenotypical manifestations of MRD21 consisting on prominent forehead, bushy and arched eyebrows, long philtrum and thin upper lip. B (a) Hematoxylin eosin staining at 20 × magnification revealing mesenchymal, epithelial and blastomatous components in the tumor. B (b) Positive immunostaining for WT1 showing a major nuclear localization.
At 26 months old, magnetic resonance imaging revealed a solid right kidney nodule in the lower pole (4 × 4.1 cm) and an additional focal lesion in the left kidney (1.9 cm maximum diameter), suggesting a bilateral WT. Chemotherapy treatment was given according to Umbrella SIOP-RTSG-2016 protocol [12] followed by a nephron sparing surgery. The right kidney lesion displayed neoplastic proliferation with blastemal and epithelial components observed in similar amounts (Fig. 1B). No signs of anaplasia were observed and, no tumor infiltration was present in the resected margin. Nuclear WT1 expression was observed by immunohistochemistry, reinforcing WT diagnosis (Fig. 1B). The left kidney lesion corresponded to a nephrogenic neoplastic nodule with nephroblastomatous-like characteristics, consisting on blastema component with considerable mitotic activity without signs of anaplasia.
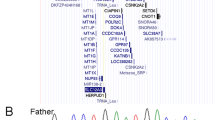
No pathogenic single nucleotide exclusively somatic variants were detected by NGS. The likely pathogenic variant CTCF c.1337 T > A (p.I446K) (exon 7, NM_006565.4) was detected heterozygous in the germline (VAF 51%) and homozygous in the tumor (VAF 97%; 659x). The homozygous detection of the CTCF variant was further confirmed in expressed tumor RNA (Fig. 2D). The variant was confirmed to be de novo in the patient after a family segregation study (Fig. 2A–C). We next used NGS results to compare CNVs within CTCF exons between tumor and blood DNA (green and red lines respectively, Fig. 2E). Despite the noisiness of the technique, no significant variations were detected (yellow line, Fig. 2E) suggesting that there were no internal deletions, gains or exome alterations aside of the single nucleotide variant found.
Sequencing alignment of CTCF variant c.1337 T > A (p.I446K) in (A) father’s germinal DNA, (B) mother´s germinal DNA, (C) patient’s germinal DNA and (D) patient’s tumor cDNA. E Copy number variations (CNV) along CTCF exons (indicated in the bottom of each segmented area) derived from NGS sequencing data from germline DNA (green line) and tumor DNA (red line). Differences between germline and tumor CNV defined by the yellow line reveal no internal CNV alterations in CTCF’s coding sequence in the tumor. F Molecular karyotype in circus-plot of the reported Wilms tumor. Allele peaks (inner plots) and weighted Log2ratio (middle plots) information were obtained from Affymetrix software ChAS. The only alteration detected was an LOH in the whole 16q arm (where CTCF gene is located).
We performed a pan-genomic analysis of the tumor in order to confirm CTCF status as well as to identify other genetic alterations. Notably, SNPa data didn´t reveal neither numerical nor segmental chromosomal alterations in the tumor (Fig. 2F). The only alteration was an LOH covering the whole q arm of chromosome 16, including CTCF gene. Further microsatellite analysis revealed that de novo mutation occurred in the mother allele.
In accordance with the first international expert consensus statement in BWS [12], the BWSp diagnosis was assessed in our patient, who would obtain a final score of three points. DNA methylation testing for both 11p15.5 imprinting control centers H19/IGF2:IG DMR (IC1) and KCNQ1OT1:TSS DMR (IC2) was indicated; the result was negative. CDKN1C loss of function mutations were ruled out as well. Considering these molecular results, the patient´s clinical features were consistent with MRD21.
Discussion
MRD21 was firstly reported by Gregor et al. in 2013 [6] and afterwards characterized in the largest series [9]. To our knowledge, this is the first diagnosed MRD21 patient that suffers from WT. Considering the rarity of bilateral WT among children, the oddity of MRD21, and the data here reported, we hypothesize CTCF variant c.1337 T > A (p.I446K) as a link between MRD21 and WT predisposition. In fact, CTCF mutations within zinc-finger domains have been described among WT patients [12]. One of two variants reported by Filippova et al. among WT samples is located within zinc-finger domain 7 (R448Q). In the presence of this variant, they describe CTCF failure to bind the Igf2/H19 sites [13]. The variant detected with a 100% allelic frequency in the tumor of our patient, located as well within zinc-finger 7, might be responsible of analogous effects. This result suggests that during tumor evolution 16q-wild-type arm was replaced by a copy of the 16q arm containing the CTCF variant, thus resulting in an LOH. Although the link between MRD21 and the observed bilateral WT is credible, further molecular assays are required to demonstrate the differential binding of CTCF variant in the Igf2/H19 sites. Importantly, binding may be conditioned by other relevant factors such as transcriptional coactivators and corepressors expressed exclusively in cells where a phenotype is observed: kidney and neurons. Thus, assays may also require the right environment to demonstrate that CTCF variant binds differentially to Igf2/H19 sites and therefore, animal models may also be required.
CTCF mutation was found de novo in the child, suggesting that the mutation may have originated in the gametes. After performing microsatellite analysis, we determined that the LOH allele in which the mutation is found corresponds to the mother, excluding its connection with the seminoma suffered by the father. Alternatively, the mother may have the mutation in a mosaicism that was not detected in blood cells by Sanger sequencing, which does not exclude of being de novo in the child because it shows now in all his cells, independently on whether there was a mosaicism in the mother.
LOH at 16q occurs in nearly 20% of WT patients and is an independent prognostic factor among low histological stage tumors [14]. 16q LOH was reported to be associated with loss of imprinting (LOI) at 11p15 and CTCF reduced expression in a group of patients [15]. Whereas CTCF gene is located at 16q22 and is a basic element in normal imprinting at 11p15, its haploinsufficiency might be responsible for 11p LOI and therefore, it may explain a driver mechanism in some WT [15]; however, more data are needed. Moreover, hypermethylation of a CTCF binding site downstream of the WT1 gene promoter would disturb the normal transcriptional regulation of WT1 and it might be considered oncogenic in WT [16]. Although the mechanisms are only partially known, CTCF is currently thought to play a role in regulating WT1 gene expression in WT. However, these data are not sufficient to demonstrate a driver role for these alterations. In fact, Cresswell GD et al. [4], supported that 16q is a heterogeneous event in WT that is unlikely to be a driver.
Mild forms of BWS have been described without fitting classic criteria [17] but, the phenotype in our patient is clearly more compatible with MRD21 than with BWSp. However, the patient was excluded from BWSp diagnosis, based on current recommendations [18]. In this case, the presence of the variant in germline would justify the development of MRD21.
Thus, the results here reported only suggest an implication in tumor development of 16q LOH carrying a CTCF likely pathogenic variant and, a possible predisposition to WT development in patients with MRD21.
Data availability
The datasets generated during and/or analyzed during the current study are available in the GEO database repository, GSE193235.
References
Charlton J, Irtan S, Bergeron C, Pritchard-Jones K. Bilateral Wilms tumour: a review of clinical and molecular features. Expert Rev Mol Med. 2017;19:1–13.
Phelps HM, Kaviany S, Borinstein SC, Lovvorn HN 3rd. Biological drivers of Wilms tumor prognosis and treatment. Child (basel). 2018;5:1–13.
Singh P, Lee DH, Szabo PE. More than insulator: multiple roles of CTCF at the H19-Igf2 imprinted domain. Front Genet. 2012;3:1–9.
Cresswell GD, Apps JR, Chagtai T, Mifsud B, Bentley CC, Maschietto M, et al. Intra-tumor genetic heterogeneity in Wilms tumor: clonal evolution and clinical implications. EBioMedicine. 2016;9:120–9.
Scott RH, Stiller CA, Walker L, Rahman N. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet. 2006;43:705–15.
Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, et al. De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet. 2013;93:124–31.
Bastaki F, Nair P, Mohamed M, Malik EM, Helmi M, Al-Ali MT, et al. Identification of a novel CTCF mutation responsible for syndromic intellectual disability - a case report. BMC Med Genet. 2017;18:1–6.
Chen F, Yuan H, Wu W, Chen S, Yang Q, Wang J, et al. Three additional de novo CTCF mutations in Chinese patients help to define an emerging neurodevelopmental disorder. Am J Med Genet C Semin Med Genet. 2019;181:218–25.
Konrad EDH, Nardini N, Caliebe A, Nagel I, Young D, Horvath G, et al. CTCF variants in 39 individuals with a variable neurodevelopmental disorder broaden the mutational and clinical spectrum. Genet Med. 2019;21:2723–33.
Moore JM, Rabaia NA, Smith LE, Fagerlie S, Gurley K, Loukinov D, et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PloS ONE. 2012;7:1–10.
Sanmartin E, Munoz L, Piqueras M, Sirerol JA, Berlanga P, Cañete A, et al. Deletion of 11q in Neuroblastomas Drives Sensitivity to PARP Inhibition. Clin Cancer Res. 2017;23:6875–87.
van den Heuvel-Eibrink MM, Hol JA, Pritchard-Jones K, van Tinteren H, Furtwängler R, Verschuur AC, et al. Position paper: Rationale for the treatment of Wilms tumour in the UMBRELLA SIOP-RTSG 2016 protocol. Nat Rev Urol. 2017;14:743–752.
Filippova G, Qi CF, Ulmer J, Moore JM, Ward MD, Hu YJ, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52.
Grundy PE, Breslow NE, Li S, Perlman E, Beckwith JB, Ritchey ML, et al. Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: a report from the National Wilms Tumor Study Group. J Clin Oncol. 2005;23:7312–21.
Mummert SK, Lobanenkov VA, Feinberg AP. Association of chromosome arm 16q loss with loss of imprinting of insulin-like growth factor-II in Wilms tumor. Genes Chromosomes Cancer. 2005;43:155–61.
Zitzmann F, Mayr D, Berger M, Stehr M, von Schweinitz D, Kappler R, et al. Frequent hypermethylation of a CTCF binding site influences Wilms tumor 1 expression in Wilms tumors. Oncol Rep. 2014;31:1871–6.
Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2010;18:8–14.
Brioude F, Kalish JM, Mussa A, Foster AC, Bliek J, Ferrero GB, et al. Expert consensus document: Clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: an international consensus statement. Nat Rev Endocrinol. 2018;14:229–49.
Funding
To Désirée Ramal for her administrative support. PG was supported by a post-resident fellowship from IIS La Fe.
Author information
Authors and Affiliations
Contributions
PG, SO, FM, MT, JB, AJ-R, AC, VC and JFM were responsible for designing the study, conducting the search and interpreting results. YY, FM, SD, IC, FM, VS, MLL performed and analyzed one or more tests included in this manuscript. PG wrote the first version of the manuscript. All authors provided feedback on the report.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The procedures employed were reviewed and approved by CEIm La Fe (February 21Th 192 2018; register number 2017/0546). Informed consent was obtained from parents of the patient.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gargallo, P., Oltra, S., Tasso, M. et al. Germline variant in Ctcf links mental retardation to Wilms tumor predisposition. Eur J Hum Genet 30, 1288–1291 (2022). https://doi.org/10.1038/s41431-022-01105-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-022-01105-x
This article is cited by
-
Genome sequencingâdo you know what you are getting into?
European Journal of Human Genetics (2022)