Abstract
Kabuki syndrome (KS) is a rare genetic disorder caused by mutations in two major genes, KMT2D and KDM6A, that are responsible for Kabuki syndrome 1 (KS1, OMIM147920) and Kabuki syndrome 2 (KS2, OMIM300867), respectively. We lack a description of clinical signs to distinguish KS1 and KS2. We used facial morphology analysis to detect any facial morphological differences between the two KS types. We used a facial-recognition algorithm to explore any facial morphologic differences between the two types of KS. We compared several image series of KS1 and KS2 individuals, then compared images of those of Caucasian origin only (12 individuals for each gene) because this was the main ethnicity in this series. We also collected 32 images from the literature to amass a large series. We externally validated results obtained by the algorithm with evaluations by trained clinical geneticists using the same set of pictures. Use of the algorithm revealed a statistically significant difference between each group for our series of images, demonstrating a different facial morphotype between KS1 and KS2 individuals (mean area under the receiver operating characteristic curve = 0.85 [p = 0.027] between KS1 and KS2). The algorithm was better at discriminating between the two types of KS with images from our series than those from the literature (p = 0.0007). Clinical geneticists trained to distinguished KS1 and KS2 significantly recognised a unique facial morphotype, which validated algorithm findings (p = 1.6e−11). Our deep-neural-network-driven facial-recognition algorithm can reveal specific composite gestalt images for KS1 and KS2 individuals.
Similar content being viewed by others
Introduction
Kabuki syndrome (KS) is a rare genetic disorder characterised by mild to moderate intellectual disability (ID), skeletal dysplasia, visceral malformations, immunopathological manifestations, dermatoglyphic anomalies including persistence of foetal fingertip pads, and specific facial features. Two major genes are responsible for the two subtypes of KS—KS type 1 (KS1 OMIM147920) and KS type 2 (OMIM300867). Pathogenic variants in KMT2D (OMIM602113 [1]) lead to KS1 and account for 34–76% of KS (autosomal dominant), whereas pathogenic variants in KDM6A (OMIM300128 [2, 3]) lead to KS2 and account for less than 10% of all KS (X-linked) [4]. Currently we lack a description of how to differentiate the subtypes of KS based on clinical phenotypic features alone.
Recent advances in high-throughput sequencing technologies have changed the strategy and process for a precise diagnosis in individuals. Before next-generation sequencing (NGS), the classical strategy was based on the ability of the clinician to identify a genetic disease by the individual’s phenotype (phenotype-first strategy). This strategy was limited to clinically recognisable phenotypes according to the facial gestalt recognition expertise of the clinical geneticist and was responsible for some delayed diagnoses. NGS provides high diagnostic yields and allows for molecular studies without a preconceived hypothesis (genotype-first strategy), a more effective approach than the phenotype-first strategy [5].
However, physicians now have difficulty identifying the narrow set of candidate variants among all examined genes and their proper interpretation. The reverse phenotyping step helps associate the candidate genotype with the observed phenotype. Nevertheless, reverse phenotyping is time-consuming and still depends on the availability of a small number of experts.
In addition to technological progress in DNA sequencing, further advances have been made with deep learning technologies in genomics, such as automated recognition of the facial phenotype [6]. Automated association of facial phenotype with a genetic disease, also called next-generation phenotyping, could allow for dissemination of facial gestalt recognition expertise of clinical geneticists during the reverse phenotyping step [7,8,9].
Automated facial recognition has been helpful in medical diagnosis [10, 11]. Computer-assisted syndromology seems even more precise than clinical geneticists’ expertise, except surprisingly for KS [12]. However, whether automated recognition can highlight specific facial features yet undescribed by humans is unknown. In this study, we used facial morphology analysis to detect any facial morphological differences between the two KS types.
Materials and methods
Image series
As the basis for this study, frontal images of individuals with a definite molecular diagnosis of KS1 or KS2 were used to capture the facial gestalt of KS individuals. A first set of pictures was collected by collaboration, and a second set was collected from the literature. References of pictures from the literature are in Supplementary Tables S1 and S2 [13,14,15,16,17,18,19]. All facial images were fully de-identified by using DeepGestalt facial analysis. For this study, we compared two sets of images: KS1 and KS2.
Statistical analysis
Capabilities of DeepGestalt technology and statistical analysis
DeepGestalt technology combines facial-recognition software with clinical knowledge (feature annotation and anthropometric measurements), thus enabling detection of dysmorphic features and recognisable patterns of human malformations from 2D facial photographs.
Process of image analysis
The photo or input image was first pre-processed for facial detection, landmark detection, and alignment. Then the input image was cropped into facial regions. Each region was fed into a Deep Convolutional Neural Network (DCNN) to obtain a softmax vector indicating its correspondence to each syndrome in the model. The output vectors of all regional DCNNs were then aggregated and sorted to obtain a final ranked list of genetic syndromes—the 30 syndrome matches displayed in the Face2Gene online tool (FDNA Inc. Boston, MA, USA). The analysis of visual facial data is used to form a mathematical representation of the face (facial descriptor), which can be readily compared to other such descriptors.
Masks/composite images and heatmaps
The facial descriptor can also be graphically displayed as a 2D model of the face specific to the particular condition of interest (mask/composite image). These 2D mask/composite images can be used to visually represent the comparison of two digital facial descriptors, and a graphical heatmap can be applied to visualise the degree of similarity between the two descriptors being compared.
Binary comparisons (receiver operating characteristic curve [ROC] and area under the ROC curve [AUC])
The comparison and separation quality between the two KS groups was evaluated by measuring the AUC. To estimate the statistical power of DeepGestalt in distinguishing KS individuals from controls, a cross-validation scheme was used, including a series of binary comparisons between all groups with the data split randomly multiple times into training and test sets. Each set contained half of the samples for the group, and this random process was repeated 10 times. The results of the binary comparisons are reported both numerically and graphically.
Comparing image size
We compared the mean file size of image from our series to those from the literature by using Student t test with the biostaTGV website (https://biostatgv.sentiweb.fr/).
Identifying facial morphology features
To compare the facial morphology of KS1 and KS2, we used the computed specific gestalts resulting from the algorithm. The gestalts were analysed by two independent experts in dysmorphology, then notable differences were translated into human phenotype ontology (HPO) terms on the HPO website (https://hpo.jax.org/).
External validation with clinical geneticists
To determine whether clinical geneticists can differentiate KS1 and KS2 facial morphology, we organised a 20-min online challenge as follows: first, we explained the goals of the study and asked participants to estimate their expertise in KS dysmorphology by ranking themselves from 1 (no expertise) to 5 (experts in KS dysmorphology). Next, we showed the clinicians slides from the literature of individuals with KS1 and KS2, authenticated by molecular genetic analysis, to allow them to identify phenotypic differences between the two subtypes.
Finally, we showed the same set of images obtained by the DeepGestalt technology algorithm. The clinicians could observe the image for 10 s and were asked to classify the images as KS1 and KS2. However, we used only 33/34 images from the set because we did not receive consent to use the image for one individual with KS2.
Statistical analysis involved using Python 3.7 and the SciPy 1.5.4 package. Shapiro test and QQ plots were used to assess the normal distribution of scores from the performance challenge. Characteristics of KS1 and KS2 groups in images were compared by Fisher exact test. One-sample Student t test was used to determine whether health professionals could distinguish a specific gestalt (more than a random distribution). Two-sided Student t test was used to assess whether self-estimated expertise affected performance.
Results
Building the datasets
Collaborative dataset
The first dataset was a national collaboration series that contained frontal facial views of 17 KS individuals (10 males) with KMT2D pathogenic variants (KS1) and 17 (10 males) with KDM6A variants (KS2). Most of the individuals were children, with only one adult in the KDM6A group and three adults in the KMT2D group. The KS1 and KS2 groups did not differ by sex, ethnicity, or genetic variant type (Supplementary Table S3).
Collaborative dataset—only individuals of Caucasian origin
The second dataset included individuals of only Caucasian origin from the national collaboration dataset: 12 with a KMT2D variant, and 12 with a KDM6A variant.
Literature dataset
This third dataset contained frontal facial views of KS individuals of different ethnic origins reported in the literature: 16 with a KMT2D variant (4 males) and 16 with a KDM6A variant (3 males) (Supplementary Table S4).
Full mixed dataset
The full mixed dataset contained the full collaborative and literature datasets (33 males and 33 females). Detailed information for age, sex, and ethnicity is in Supplementary Table S5).
Full mixed dataset—only Caucasian origin
This dataset contained images of 49 KS individuals (23 with a KDM6A variant and 26 with a KMT2D variant) of Caucasian origin from the national collaboration and literature datasets.
Statistical analysis
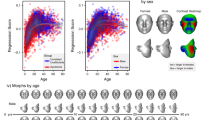
The AUC was used to measure the comparison and separation quality between the KS1 and KS2 groups. For the collaborative dataset, the AUC was 0.837 (p = 0.027) (Fig. 1A). The mean AUC was 0.85 with SD 0.08 (Supplementary Table S6).
For the second dataset, the AUC was 0.868 (p = 0.028) (Fig. 1B). The mean AUC was 0.89 and SD 0.05 (Supplementary Table S7).
For the full mixed dataset, the AUC was 0.722 (p = 0.022) (Fig. 2A). The mean AUC was 0.74 and SD 0.04 (Supplementary Table S8).
For the full mixed dataset with only Caucasian individuals, the AUC was 0.752 (p = 0.029) (Fig. 2B). The mean AUC was 0.77 and SD 0.05 (Supplementary Table S9).
Computed KS1- and KS2-specific gestalt
The specific gestalt composites for KS1 and KS2 carrying KMT2D or KDM6A variants, respectively, computed from collaborative dataset (17 individuals per KS group), are in Fig. 3. In terms of the whole KS composite gestalt image, the main morphological differences were located on the midface shape. Individuals with KMT2D variants seemed to have a longer face (HPO no.: HP:0000276), longer nose (HP:0003189), thin upper lip vermilion (HP:0000219) and longer middle part of the face (no HP number) than KDM6A variant individuals, whereas individuals with KDM6A variants seemed to share a round face (HP:0000311), thick vermilion border (HP:0012471) and anteverted nares (HP:0000463)
Comparing mean file size
The mean picture file size was 901 Ko and 133 Ko for the collaborative and literature datasets, respectively (p = 0.0007).
Clinicians’ control experiment
We recruited 60 clinical geneticists (Supplementary Table 10), with 2 groups of KS facial morphology expertise: (1) 28 with an expertise score of 1 or 2 (1–2 group) and (2) 32 with a score of 3 or 4 (3–4 group), with higher expertise. The score performance for clinicians was normally distributed (Shapiro test p = 0.14, Supplementary Fig. S10). After the training session, the overall cohort of clinicians significantly recognised a specific gestalt between KS1 and KS2 individuals (mean score = 20/33 correctly assessed images, one-sample Student t test p = 1.60e−11). The performance significantly differed by self-evaluated expertise in KS (two-sided Student t test p = 6.49e−4, Fig. 4). Each subgroup of expertise significantly distinguished a specific gestalt (1–2 group: mean 18.5, one-sample Student t test p = 2.23e−3; 3–4 group: mean 21.28, one-sample Student t test p = 1.04e−10). All statistics are in Supplementary Table S11.
Discussion
The results of our algorithm using the collaborative dataset of images show a statistically significant difference between the KS1 and KS2 group images strongly suggesting a different facial morphotype, in particular regarding the midface shape (nose, philtrum, and upper lips). The removal of individuals of African origin had a slight positive effect on the AUC, although not significant, which implies that the phenotypes of KS1 and KS2 are probably still recognisable by the Face2Gene tool for individuals of African origin.
For the literature dataset, performance of the algorithm decreased greatly. One of the parameters that could explain this difference is image quality. Our collaborative dataset overall had very good image quality, whereas the literature images were usually of poor quality because they were taken as screenshots. Thus, good image quality seems important for the software to correctly assess and compare the images.
Collecting additional good-quality images from KS1 and KS2 individuals could help better decipher facial features associated with the genotype. Moreover, because of the lack of individuals of Asian origin in our collaborative dataset, we cannot conclude on the algorithm’s ability to distinguish between KS types among individuals of Asian ethnicity.
Studying a larger series with more ethnicities would definitely be a great step to better delineate the facial phenotype between KS1 and KS2.
We also tried to determine whether humans could differentiate between KS1 and KS2 phenotypes. Clinicians with basic training in dysmorphology could indeed distinguish between the two KS groups. Clinicians with better self-reported dysmorphology expertise performed better than those with self-reported basic expertise. Thus, with training, human geneticists could distinguish phenotypes, and even better for those specialised in KS dysmorphology.
As in previous studies, these results support the significant discrimination capacity of a deep-neural-network-driven facial-recognition algorithm that may imply the possibility of using next-generation phenotyping in the diagnostic process of individuals with ID as long as images of good graphic quality are used.
Here we report a specific gestalt for KS1 and KS2 individuals revealed by use of a deep-neural-network-driven facial-recognition algorithm. Because the genetic determinants of craniofacial development are continuously being discovered, a precise facial phenotypic description could help in the biological interpretation of variants from whole exome and genome sequencing [20, 21]. Artificial intelligence technologies could push the boundaries of phenotype recognition beyond human capacity for identifying additional facial features of syndromic genetic disorders.
Data availability
The data (patient’s facial pictures) that supports the findings of this study are available from the French research program PHRC AOM-09-070 (ClinicalTrials. gov identifier: NCT01314534), but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of patients and physicians in charge of the patients.
References
Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3.
Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, et al. Deletion of KDM6A, a Histone Demethylase Interacting with MLL2, in Three Patients with Kabuki Syndrome. Am J Hum Genet. 2012;90:119–24.
Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34:108–10.
Bögershausen N, Gatinois V, Riehmer V, Kayserili H, Becker J, Thoenes M, et al. Mutation Update for Kabuki Syndrome Genes KMT2D and KDM6A and Further Delineation of X-Linked Kabuki Syndrome Subtype 2. Hum Mutat. 2016;37:847–64.
Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BWM, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7.
Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. A primer on deep learning in genomics. Nat Genet. 2018. https://doi.org/10.1038/s41588-018-0295-5
Liehr T, Acquarola N, Pyle K, St-Pierre S, Rinholm M, Bar O, et al. Next generation phenotyping in Emanuel and Pallister-Killian syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin Genet. 2018;93:378–81.
Basel-Vanagaite L, Wolf L, Orin M, Larizza L, Gervasini C, Krantz ID, et al. Recognition of the Cornelia de Lange syndrome phenotype with facial dysmorphology novel analysis. Clin Genet. 2016;89:557–63.
Hadj-Rabia S, Schneider H, Navarro E, Klein O, Kirby N, Huttner K, et al. Automatic recognition of the XLHED phenotype from facial images. Am J Med Genet A. 2017;173:2408–14.
Hurst ACE. Facial recognition software in clinical dysmorphology. Curr Opin Pediatr. 2018;30:701–6.
Pantel JT, Zhao M, Mensah MA, Hajjir N, Hsieh T-C, Hanani Y, et al. Advances in computer-assisted syndrome recognition by the example of inborn errors of metabolism. J Inherit Metab Dis. 2018;41:533–9.
Dudding-Byth T, Baxter A, Holliday EG, Hackett A, O’Donnell S, White SM, et al. Computer face-matching technology using two-dimensional photographs accurately matches the facial gestalt of unrelated individuals with the same syndromic form of intellectual disability. BMC Biotechnol. 2017;17:90.
Moon J-E, Lee S-J, Ko CW. A de novo KMT2D mutation in a girl with Kabukisyndrome associated with endocrine symptoms: a case report. BMC Med Genet. 2018;19:102.
Li Y, Bögershausen N, Alanay Y, Simsek Kiper PO, Plume N, Keupp K, et al. A mutation screen in patients with Kabuki syndrome. Hum Genet déc. 2011;130:715–24.
Mısırlıgil M, Yıldız Y, Akın O, Odabaşı Güneş S, Arslan M, Ünay B. A Rare Cause of Hyperinsulinemic Hypoglycemia: Kabuki Syndrome. J Clin Res Pediatr Endocrinol. 2020. Online ahead of print.
Lederer D, Shears D, Benoit V, Verellen-Dumoulin C, Maystadt I. A three generation X-linked family with Kabuki syndrome phenotype and a frameshift mutation in KDM6A. Am J Med Genet A. 2014;164A:1289–92.
So PL, Luk HM, Yu KPT, Cheng SSW, Hau EWL, Ho SKL, et al. Clinical and molecular characterization study of Chinese Kabuki syndrome in Hong Kong. Am J Med Genet A. 2021;185:675–86.
Dentici ML, Di Pede A, Lepri FR, Gnazzo M, Lombardi MH, Auriti C, et al. Kabuki syndrome: clinical and molecular diagnosis in the first year of life. Arch Dis Child. 2015;100:158–64.
Banka S, Lederer D, Benoit V, Jenkins E, Howard E, Bunstone S, et al. Novel KDM6A (UTX) mutations and a clinical and molecular review of the X-linked Kabuki syndrome (KS2). Clin Genet. 2015;87:252–8.
Gurovich Y, Hanani Y, Bar O, Nadav G, Fleischer N, Gelbman D, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25:60–4.
Van der Donk R, Jansen S, Schuurs-Hoeijmakers JHM, Koolen DA, Goltstein LCMJ, Hoischen A, et al. Next-generation phenotyping using computer vision algorithms in rare genomic neurodevelopmental disorders. Genet Med. 2018. https://doi.org/10.1038/s41436-018-0404-y
Acknowledgements
We deeply thank all clinicians and biologists involved in diagnostic and data sharing for this study. We thank the French Kabuki Association for their help for this study. We thank Nicole Fleischer and Sarah Savage for advice and assistance related to the algorithm for this project.
Funding
Part of this work was supported by the French Ministry of Health (Programme Hospitalier de Recherche Clinique national, AOM 07-090), Fondation Maladies Rares, and the French Kabuki Association http://www.syndromekabuki.fr/.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
DG was a consultant for the Takeda Society in 2018. Takeda did not have any role in this study.
Ethical approval
All clinical geneticist consents for participation were obtained through a survey where their responses were also collected.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Rouxel, F., Yauy, K., Boursier, G. et al. Using deep-neural-network-driven facial recognition to identify distinct Kabuki syndrome 1 and 2 gestalt. Eur J Hum Genet 30, 682–686 (2022). https://doi.org/10.1038/s41431-021-00994-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-021-00994-8
This article is cited by
-
Next generation phenotyping for diagnosis and phenotype–genotype correlations in Kabuki syndrome
Scientific Reports (2024)
-
2022: the year that was in the European Journal of Human Genetics
European Journal of Human Genetics (2023)
-
What’s new in genetics in June 2022?
European Journal of Human Genetics (2022)