Abstract
A primary challenge in understanding disease biology from genome-wide association studies (GWAS) arises from the inability to directly implicate causal genes from association data. Integration of multiple-omics data sources potentially provides important functional links between associated variants and candidate genes. Machine-learning is well-positioned to take advantage of a variety of such data and provide a solution for the prioritization of disease genes. Yet, classical positive-negative classifiers impose strong limitations on the gene prioritization procedure, such as a lack of reliable non-causal genes for training. Here, we developed a novel gene prioritization tool—Gene Prioritizer (GPrior). It is an ensemble of five positive-unlabeled bagging classifiers (Logistic Regression, Support Vector Machine, Random Forest, Decision Tree, Adaptive Boosting), that treats all genes of unknown relevance as an unlabeled set. GPrior selects an optimal composition of algorithms to tune the model for each specific phenotype. Altogether, GPrior fills an important niche of methods for GWAS data post-processing, significantly improving the ability to pinpoint disease genes compared to existing solutions.
Similar content being viewed by others
Introduction
Despite the tens of thousands of genetic associations identified using GWAS to date, the ultimate goal—informing and guiding therapeutic development—has been achieved for only a few phenotypes. A major complication in understanding disease biology from GWAS often arises from the inability to directly identify disease genes [1]. Therefore, additional post-GWAS analysis is needed to first, identify a variant that drives the signal within the locus, and then to connect this variant to a gene.
Fine-mapping, based on a Bayesian framework, sets out to prioritize variants within the locus and, ultimately, to identify the disease-causing variant [2,3,4]. Fine-mapping algorithms—FINEMAP [5], PAINTOR [6], fGWAS [7], SUSIE [8], etc. have made a significant impact on the field and have helped to successfully identify causal variants for multiple traits. Importantly, fine-mapping is done independently for each locus and in its current configuration does not take advantage of biological relatedness (e.g., the same pathway membership) of genes involved in a phenotype [9].
At the same time, identification of the disease-relevant gene linked to a disease-associated variant presents a major, unresolved challenge to gaining biological insight from the genetic association. Most GWAS associations implicate a set of correlated genetic variants, none of which alter the protein-coding sequence of a gene and which often physically span or are near to multiple genes. Since our knowledge of regulatory sequence patterns of the genome, as well as the cells, tissues, and developmental time-points most relevant to disease, are all incomplete, it is currently the case that the vast majority of GWAS “hits” do not have an established link to a gene—though data sets with which to infer functional annotation and gene expression are growing rapidly in their utility.
Analytic methods for post-processing GWAS results using functional information are therefore promising tools for disease gene identification. For example, Post-GWAS Analysis Platform (POSTGAP [10]) uses GWAS summary statistics along with Linkage Disequilibrium (LD) structure and external functional databases (GTEx [11], FANTOM5 [12], RegulomeDB [13]) to prioritize SNPs within the locus and narrow down the list of potential gene candidates. Yet, the gene prioritization utility of POSTGAP is still in early development and has not been fully tested.
Altogether, fine-mapping, functional annotations, and known biologic relatedness across putative disease genes can become valuable data sources for gene prioritization, which we define as the evaluation of the likelihood of a gene being causally involved in generating a disease phenotype [14]. Machine-learning (ML) based prioritization could take an advantage of these data sources and provide a solution for novel disease gene identification.
Typically, existing ML solutions use Positive-Negative (PN) classification strategy. In PN classification per-gene probabilities are obtained by using known disease genes as a positive (P) training set and unknown genes as a negative (N) training set [15,16,17,18]. In addition to often having a limited set of confirmed positives in each disease, such an approach suffers from contamination of a negative set by hidden positives (HP), represented by yet undiscovered disease genes. In addition, it is challenging to find reliable negative examples (i.e., genes that with certainty do not contribute to the development of a phenotype). Most biological databases do not store negative evidence (e.g., absence of gene interaction), rather they provide only observed positive evidence. As a result, particularly in highly polygenic traits, PN-classifiers could suffer from high false-negative prediction rates and biased quality metrics.
It is feasible to design a model, where a limited number of reliable positive examples (likely causal genes) will be used along with all remaining genes without treating the latter as reliable negatives. PU-learning treats unknown examples as a mixture of P and N, called unlabeled (U) set, and has been developed to overcome limitations of PN-learning.
A theoretical study of PU-learning was first conducted by Denis et al. [19], and several algorithms have been published since then [20, 21]. A particular class of PU methods—PU-bagging, showed the best stability of the learning algorithm. Specifically, Mordlet et al. [22] proposed the “bagging SVM” approach that took advantage of a limited number of positive examples and significantly improved the performance and stability of classification using a bootstrap aggregating technique. It treats all provided positives as TP and iteratively subsamples the same size of unlabeled instances, using them as negatives. Thus, on each iteration only a small portion of U is treated as N, minimizing false-negative error rate [22].
Nevertheless, a single ML algorithm cannot fit all complex phenotypes and highly heterogeneous biological data. To overcome this, Yang et al. [23] introduced a method that integrated several PU learning classifiers into one workflow using ensemble technique. This technique was only tested with a specific family of PU algorithms—two-step methods, heuristic in nature, and sensitive to the initial choice of negative examples [24], significantly limiting applicability to GWAS data. Two-step PU algorithms first attempted to identify negative examples in the unlabeled set, and then train a model from the positive, unlabeled, and likely negative examples. Such algorithms are preferable when classes are close to each other, but at least separable [21]. However, in most polygenic traits, heterogeneous gene-based data are far from being divisible. Directly learning to discriminate P from U with the estimation of optimal misclassification costs is preferable in this case.
Therefore, a composition of different ML algorithms along with PU bagging is a promising strategy for building a gene-prioritization model suitable for a large number of complex phenotypes and a high variety of data sources, which is still lacking in the field.
Here, we propose a novel gene prioritization tool based on PU-learning—Gene Prioritizer (GPrior), intended for post-fine-mapping interpretation of GWAS results. In GPrior we implemented the ensemble of 5 different ML classifiers for PU-bagging with a further selection of the optimal composition of predictions. Our approach returns probability scores for the provided set of genes based on similarity level with positive examples used for training. It is complementary to other gene prioritization tools and fine-mapping techniques further expanding potential usage scenarios.
We illustrate the utility of PU-learning and GPrior for the disease gene prioritization with a series of case studies and validation experiments. Comparison with popular methods (TOPPGENE [25]; Bagging SVM [22]; MAGMA [26]) confirmed significantly higher quality of predictions returned by GPrior.
Methods
GPrior was designed for prioritizing disease-relevant genes given a matrix of gene-level features and a set of reliably causal genes. We integrated multiple ML techniques in a single tool with a data-driven framework to select the most appropriate algorithm (or composition of algorithms) on a case-by-case basis.
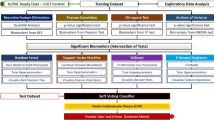
The prioritization scheme includes two independent steps. First, each ML algorithm is used for positive-unlabeled bagging and generation of predictions for each gene. Second, the best-performing composition of predictions is generated. To ensure the independence of steps, GPrior uses two independent training sets—a set of genes for training individual ML algorithms and a second algorithm evaluation set (Fig. 1, Supplementary Fig. S1). The latter is used to evaluate the quality of predictions from ML algorithms and select predictions that will contribute to the optimal composition. Altogether, such an approach allows the composition of multiple learning algorithms and can thereby achieve performance previously unattainable for an individual algorithm.
Matrix of gene features along with a vector of supervised answers is used to train 5 models (LR Logistic Regression, SVM Support Vector Machine, RF Random Forest, DT Decision Tree, AB Adaptive Boosting) using PU-bagging approach. Two independent gene sets are used for training –a true set of genes for individual classification algorithms training and algorithm evaluation set of true genes for selecting the optimal composition of the predictions. Predictions are generated using positive-unlabeled bagging and further, an optimal composition returning the largest PU-score is returned.
Input and features
In addition to the described above true gene sets needed for training, GPrior requires a data matrix with rows representing genes and columns representing features. Note that training sets were defined prior to running GPrior. To avoid sampling bias, we used three different gene set compiling strategies: GWAS Catalog mining, publication-based and expert curation (Supplementary Materials, Training and validation gene sets).
GWAS summary statistics contain only variant information that needs to be converted into gene-level data. Initially, we filtered out likely non-associated variants with the p-value threshold determined on a case-by-case basis, ensuring the inclusion of the majority of potentially causal genes into the prioritization analysis, even if no significant association was observed in GWAS. The threshold depended on the trait polygenicity and the number of already discovered signals. We used p value < 10−8 for well-studied traits, otherwise, for highly-polygenic traits, we took variants with p value < 10−6. Further, GWAS p values were not a part of the prioritization model.
Next, we used POSTGAP [10] with the default parameters to assign gene candidates for each variant (could be more than one gene)—LD threshold of r2 > 0.7 and variant functional annotations. Such preprocessing of GWAS summary statistics yields a variant-based data matrix with mappings to an extensive list of candidate genes.
A major challenge in transforming variant-based data into a gene-based data matrix for GPrior is the preservation of valuable information about variant annotations. We used a transformation of variant-level features (e.g. functional annotations, GERP scores, etc) into gene-level features using a method proposed by Lehne et al. [27] to obtain a gene-based data matrix.
In addition, we used gene expression and gene interaction data that proved their utility for the gene identification problem in previous works [15, 28, 29]. Specifically, we used the GTEx database to obtain median gene expression levels for 53 tissues, the Reactome database to obtain interaction data, UCSC Gene Sorter to obtain protein homology information, etc. (Supplementary Table S1). All of them were used as features for further gene annotation.
Additional functional features and predictions of other prioritization algorithms could be included in the data matrix to be used for the GPrior model to boost the performance quality [30]. GPrior could take as input either the raw output of POSTGAP (variant-based data matrix) or any gene-based data matrix provided by a user.
We kept the same set of features for the case studies to preserve the fairness of performance comparisons for different phenotypes. Although, for each phenotype, features could be selected in concordance with phenotype-specific needs, for example, relevant cell type expression data. Overall, GPrior is not bound to POSTGAP pre-processing or a pre-specified set of features and could be used with more advanced variant-to-gene mapping algorithms and a user-defined set of features to boost the trait-specific performance (Supplementary Methods, Input and Features).
GPrior algorithm
GPrior consists of five PU Bagging ensembles, each of them uses a different classification algorithm: Logistic Regression (LR), Support-Vector Machine (SVM), Decision Tree (DT), Random Forest (RF), Adaptive boosting (AB) (Fig. 1, Supplementary Methods, GPrior Algorithm).
Each positive-unlabeled bagging procedure starts with the creation of a training set with all positive (P) instances, treated as Positives, and a random subsample of unlabeled, size of P, treated as negatives (bN). Resulting in the total size of a bootstrap sample being equal to 2 P. However, it is possible to tune the bN:P ratio in GPrior parameters. This way, on each iteration only a small portion of unlabeled instances is treated as negatives, minimizing the false-negative error rate (Supplementary Fig. S2). Each learning method is then fine-tuned by finding an optimal set of parameters (Supplementary Table S2).
After training and tuning, individual classifiers generate a probability score for each gene to belong to the positive class. All the steps are repeated T times. Per-gene probabilities are obtained by dividing the sum of all predictions by the number of times each gene was sampled from the unlabeled set. All the predictions are averaged and stored as a final PU Bagging result. All the steps are repeated for each classification algorithm.
Next, GPrior selects the composition of predictions that shows the best performance in prioritizing true genes given by an independent algorithm evaluation set. Since “true negative” data points that falsely were classified as positives could not be identified in PU-data, any metric depending on false positives could not be applied for quality evaluation. Furthermore, separation of all genes into confident “risk” and confident “non-relevant” classes is not known for any polygenic trait. Rather we are limited only by “currently identified” risk genes, which makes usage of common classification quality metric (e.g. F1 score) inaccurate. Thus, we used PU-score as a formal quality metric suitable for positive-unlabeled data classes [31, 32].
We performed a set of experiments using public benchmarks data to confirm similar behavior (Pearson’s correlation R = 0.99, p value < 2.2e−16) of F1-score and PU-score, to justify further usage and interpretation of prediction quality assessment with PU-score (Supplementary Methods, Supplementary Figs. S5–S7).
All compositions from individual predictions are evaluated using PU-score calculated for the algorithm evaluation set and the best performing composition of methods is then selected as the best fitting for a given phenotype. The selected composition is used to return a vector of probabilities corresponding to the genes in the input matrix.
Results
The number of known true positive and negative data points is the essential information for gene prioritization. It is challenging to estimate both the number of genes involved in a complex trait and the number of genes confidently irrelevant to the disease.
Height GWAS, as a classic example of a highly polygenic trait study, shows substantial effect sizes for variants that are not reaching genome-wide significance. This suggests that significant associations are observed only for a small proportion of true positive data points, while many others are yet to be confirmed. However, additional alleles at known genes are a likely source of much of what is missing from disease-relevant variants or genes, so this does not easily translate into an estimate of how many relevant genes are implicated from a GWAS [33].
As shown in recent work, the genetic architecture of height is broadly similar to that of a wide variety of other quantitative traits and diseases ranging from diabetes and autoimmune diseases to BMI and cholesterol levels [33]—for all of which the evidence suggests many more positive genes exist in the “currently not associated” gene set. In addition, only a fraction of the genome-wide significant loci were mapped to a single gene, even further reducing the number of known true genes suitable for training a model. Therefore, it is reasonable to assume that gene prioritization algorithms should expect to be trained using only a small fraction of all disease-relevant genes.
Furthermore, in the absence of reliable sets of genes conclusively unrelated to a disease, it is unclear how to validate prioritization results based only on positive examples.
We conducted several benchmarking experiments to validate the utility of PU-learning for gene prioritization and check the applicability of some of the commonly used quality scores. First, we used a Breast Cancer Wisconsin (Diagnostic) data set, a popular public dataset for machine learning benchmarking, to compare the performance of a PU-learning approach and a conventional PN-learning method. Further, we tested the interchangeability of two quality scores (F1-score and PU-score). F1-score is immeasurable in real data experiments, thus we wanted to validate the utility of PU-score on a data set with known ground truth prior to conducting case studies.
Benchmarking confirmed the superior performance of PU-learning compared to a single PN-learning approach. Specifically, the PU-bagging algorithm is most useful when the fraction of known to-date true instances is below 20%, which is quite clearly the case for the majority of polygenic traits (Supplementary Methods, Benchmark data experiments; Supplementary Figs. S5–S7).
Further, we conducted a series of case studies using complex disease GWAS data to evaluate performance and compare GPrior with other methods in the real-life setting that the application is targeted towards.
Case study 1: inflammatory bowel disease (IBD)
We used GPrior and summary statistics from Huang et al. [34], to construct gene prioritization for IBD. It consisted of 67,852 individuals of European ancestry, including 33,595 with IBD (18,967 Crohn’s disease and 14,628 ulcerative colitis) and 34,257 healthy controls. Summary statistics were preprocessed to obtain a data matrix with 2,166 gene candidates found in loci with original p-value < 10−8. A list of 31 genes with known evidence to be likely causal for IBD was used as a positive training set [35,36,37,38]. The algorithm evaluation set consisted of 14 genes reported in monogenic loci with p value < 10−10 found in the GWAS catalog. We use “monogenic loci” here to refer to GWAS loci with either a single gene in a fine-mapped region or a coding variant in a credible set in the corresponding publication. Independent validation set used only for performance evaluation included 51 genes found within monogenic loci with p-values falling in range 10−10−10−8 (Fig. 2A, Supplementary Table S3). For more details about gene sets compiling procedure see Supplementary Materials, Training and validation gene sets.
A Scheme for selection of training, algorithm evaluation, and validation gene sets; B Classification quality comparison for GPrior, Bagging SVM, TOPPGENE and conventional PN-learning with weighted linear regression; C Cumulative gains curve shows better prioritization of true genes at the top of the candidate list using GPrior in comparison with other methods; D True genes from the independent validation gene set receive significantly higher scores than genes found within the same loci but not implicated inthe disease; E Enrichment of true genes from independent validation gene set among top predictions from GPrior.
In addition, we validated the importance of training set compiling procedure and influence of contamination on the prioritization quality (Supplementary Methods, GPrior Algorithm).
We generated gene priorities using GPrior (optimal composition: LR, RF) (Supplementary Table S4) and a set of methods for comparison—a single PU-learning (bagging-SVM [22]), PN-learning (weighted LR [31]), and TOPPGENE [25]. While GPrior implies two training steps and usage of two training gene sets—true gene set and algorithm evaluation set, for other methods we used a union of the two gene sets for training.
Next, we compared the performance quality of the methods. PU-score is a formal quality metric for an ML-based classifier, rather than for a prioritization itself, and it depends on the decision threshold used to assign classes to the instances. Gene prioritization implies only a construction of the ranked list of genes, but not the classification of the genes into “disease” and “non-disease”. Yet, we evaluated the maximal possible performance of the methods in the classification problem. We used an independent validation gene set to estimate PU-scores for all possible decision thresholds (fraction of positive predictions made by the classifier) and GPrior has a significantly greater maximal PU-score compared to others (Fig. 2B).
To evaluate prioritization quality, we estimated cumulative gains. The gain chart shows enrichment of the genes from the validation set at the top of the ranked list of predictions, that is the sharper is the growth of gain at the beginning of the chart – the more enriched are correct predictions at the top of the predictions list (Fig. 2C).
Since original GWAS summary statistics were preprocessed to include only variants with p value < 10−8, all 2166 genes in the data matrix are found in or in the proximity of significantly associated locus. GPrior does not use association strength or DNA location information for gene prioritization. Yet, genes from the validation set are significantly prioritized over the non-relevant neighbors (+/−250 kb, N = 239, Mann–Whitney, one-sided, p value = 4.335 × 10–6, Fig. 2D).
We evaluated the non-randomness of the predictions, by estimating enrichment of the validation set genes at the top of the ranked list produced by GPrior (permutation p value < 1*10–6; Fig. 2E).
Treatment of all genes from the validation set as a finite set of disease genes implies that all genes that are not in the validation set are true negatives. In the case of polygenic traits, this is most likely a false assumption that would lead to an underestimation of the true value of the area under the ROC curve (ROC AUC). Thus AUC values will illustrate only approximate quality measurement. In such settings, GPrior demonstrated the most efficient predictive power out of all tools (AUC = 0.8, Supplementary Table S5).
Case study 2: educational attainment
We performed a control experiment to demonstrate that GPrior predictions are disease-specific and are driven by underlying biological similarities for disease-related genes. We considered two phenotypes with a very modest expected overlap in underlying biological causes—IBD and educational attainment (EA). We hypothesized that usage of the training gene set fitted for IBD should fail to predict genes for EA.
GWAS summary statistics data from Lee et al. [39] for EA, which included 1,131,881 individuals, was preprocessed to obtain the feature matrix for candidate genes (N = 10,638), found in loci with original p value < 10–6. Educational attainment is a highly polygenic trait and modern genetic studies were able to explain only a relatively low proportion of phenotypic variation, thus we used the p-value threshold of 10–6 to incorporate even moderate signals for further analysis.
To eliminate potential bias in the size of training sets for the two phenotypes, we used for GPrior training only 18 genes (12 for ML training and 6 for algorithm selection) from the IBD training gene set that were also found in EA GWAS loci with p value < 10–6. As a validation set for IBD we used the original IBD validation genes (N = 51), for EA we used 381 genes found in monogenic loci from GWAS catalog EA results (Supplementary Table S6, Supplementary Methods).
Usage of appropriate training set for IBD resulted in significant enrichment (permutation P < 10–6) of validation set genes in the top predicted genes (Supplementary Fig. S8A). Predictions based on the same list of training genes were constructed for EA and demonstrated no enrichment of the EA-specific validation gene set (permutation p value = 0.12, Supplementary Fig. S8B). Yet, usage of the EA-specific training gene set of the same size (N = 18, Supplementary Methods) led to successful prioritization of EA-specific genes (permutation P < 10–6, Supplementary Fig. S8C).
We expanded the training set for EA by including all genes found in monogenic loci in GWAS Catalog with (N = 119) and repeated prioritization analysis. As a result, we obtained even more significant enrichment for the validation set and confirmed superior performance quality of GPrior (optimal composition: LR, Ada, RF) in comparison with other methods (Supplementary Fig. S9, Supplementary Tables S8, S9). Genes from the validation set had significantly higher probability scores in comparison with neighboring genes (+/−250 kb, N = 1390, Mann–Whitney, one-sided, p value < 2.2e-16).
Further, we utilized a set of previously undescribed genes associated with developmental disorders from a recent Kaplanis et al. [40] study to validate GPrior prioritization results. 183 of them were overlapped with our dataset and did not occur in other previously used gene sets, thus we used them as an additional validation set. GPrior successfully achieved significant prioritization of these genes (permutation p value = 2 × 10–5). Some of them CAMTA1, ZEB2, CAMK2A, TUBB3, NFIA, SHANK3, WAC, PAFAH1B1, UBTF, NFIX, MEF2C, and CAMK2B were placed at the top 200 genes.
We estimated how strongly GWAS summary statistics preprocessing with POSTGAP contributed to the overall success of the prioritization. POSTGAP, which was used for mapping variants to an expanded list of candidate genes, was not originally designed for gene prioritization. The package reports a variant-to-gene (V2G) mapping score based on the sum of values for 7 features that potentially could be used for ranking genes (Supplementary Fig. S10). We used the maximal variant-to-gene score for each gene to construct a ranked list of genes (score_max). Next, we estimated the largest possible PU-score using only POSTGAP-based gene ranking for educational attainment data (PU-score = 3.82). To evaluate the advancement in prediction quality due to a model design we limited feature space to exactly the same 7 features and ran gene prioritization using GPrior, which resulted in a nearly 10% increase in PU-score (PU-score = 4.1).
POSTGAP score is limited to the initial 7 features used to construct score_max. In turn, GPrior can take advantage of all available feature space. Including all available features into the GPrior model yielded a significant increase in quality (~27%) leading to the maximal PU-score of 4.84 (Supplementary Fig. S10).
Case study 3: Coronary artery disease (CAD)
We used the summary statistics of coronary artery disease GWAS of 34,541 CAD cases and 261,984 controls from UK Biobank followed by replication in 88,192 cases and 162,544 controls [41]. After preprocessing we obtained a gene-based data matrix with 2,794 gene candidates found in loci with original p value < 10−8.
A recent review by Khera and Kathiresan [42] was used to compile gene sets for GPrior (Fig. 3A). All genes with identified biological roles in any of the known disease pathways were used for the training set (TS = 18, AES = 8). All other genes, implicated in CAD and mentioned in the review, were used as validation set (VS = 37) (Supplementary Table S9).
A Scheme for selection of training, algorithm evaluation, and validation gene sets; B Classification quality comparison for GPrior, Bagging SVM, TOPPGENE and conventional PN-learning with weighted linear regression; C Cumulative gains curve shows better prioritization of true genes at the top of the candidate list using GPrior in comparisonwith other methods; D True genes from the independent validation gene set receive significantly higher scores than genes found within the same loci but not implicated inthe disease; E Enrichment of true genes from independent validation gene set among top predictions from GPrior.
Prioritization list obtained with GPrior (optimal composition: Ada, RF) (Supplementary Table S10) has shown the best accuracy with all quality metrics in comparison with other methods (Fig. 3B–E, Supplementary Table S11). GPrior showed both significant enrichment of genes from VS at the top of the prioritized geneset (p value < 10–6) and significantly higher probability scores in comparison with neighboring genes (+/−250 kb, N = 100, Mann–Whitney, one-sided, p value = 2.973 × 10–7).
Conclusively, using risk genes with known molecular pathway membership GPrior successfully prioritizes genes with yet unknown biological contribution but confidently implicated in the disease. Importantly, by further analyzing feature importance in the prediction model it is possible to build testable biological hypotheses for novel genes discovered in predictions.
Case study 4: Schizophrenia
We used GWAS Summary statistics from Pardiñas et al. [43]. This study used genotypes of 105,318 individuals—40,675 schizophrenia cases and 64,643 controls.
After preprocessing we obtained a gene-based data matrix with 3831 gene candidates found in loci with original p value < 10−6.
The training set was prepared using reported genes found in monogenic loci from GWAS meta-analysis results [43]. Training gene set for individual ML algorithms included 20 genes with p values falling in the range 10–44–10–14, and algorithm evaluation set included 24 genes with p values within 10−13–10−8 range. The validation set (VS) included 28 genes and was obtained from the same study and included all genes from significant polygenic loci (Fig. 4A, Supplementary Tables S12 and S13).
A Scheme for selection of training, algorithm evaluation, and validation gene sets; B Classification quality comparison for GPrior, Bagging SVM, TOPPGENE and conventional PN-learning with weighted linear regression; C Cumulative gains curve shows better prioritization of true genes at the top of the candidate list using GPrior in comparison with other methods; D True genes from the independent validation gene set receive significantly higher scores than genes found within the same loci but not implicated in the disease; E Enrichment of true genes from independent validation gene set among top predictions from GPrior; F Scheme describing the selection of additional validation set from Singh et al. publication; G Enrichment of genes from Singh et al. validation set; H Scheme describing the selection of additional validation set from PGC3; I Enrichment of genes from the PGC3 validation set.
GPrior (optimal composition: SVM, DT, RF) demonstrated superior results in comparison with other methods using all quality metrics. GPrior achieved the highest PU score (9.64) and AUC (0.92) values. On all the top intervals of the predictions list (1%, 5%, 15%, 25%) GPrior showed the highest enrichment of the validation set genes (Fig. 4B–E, Supplementary Table S14). Also, the latter had significantly higher probability scores in comparison with neighboring genes (+/−250 kb, N = 162, Mann–Whitney, one-sided, p value = 5.202 × 10−6).
Further, we compiled two additional validation sets to evaluate the disease-specific nature of the GPrior prediction and test the ability of the proposed method to predict disease genes from the latest schizophrenia studies based on the previously published data. The first set was derived from Singh et al. [44] analysis of schizophrenia exomes. We used 10 genes identified with FDR < 5% as associated with the disease that overlapped with the initial set of genes from the feature table and used them as a validation set (Fig. 4F; Supplementary Table S12). GPrior successfully achieved significant prioritization of these genes (permutation p value = 0.00356; Fig. 4G), confirming the overlap between associations for schizophrenia identified in GWAS and in rare variant studies. Several known schizophrenia-associated genes from this list were placed in the top 200 hits: GRIN2A, MAGI2, SP4, and STAG1 [45,46,47,48].
The second validation set was derived from the recently published data of Psychiatric Genomics Consortium (PGC3) [48]. We took 16 genes from monogenic loci that weren’t overlapping with genes previously used for compiling TS, AES or any VSs (Fig. 4H; Supplementary Table S12). GPrior demonstrated even more superior enrichment (p value < 10−6; Fig. 4I). 5 genes from the list were presented in the top 200 hits: CALN1, NEGR1, NMUR2, WSCD2, PPARGC1A. Additional trait-specific feature selection and integration of both common and rare variants data into the analysis can further significantly improve the overall quality of schizophrenia disease genes prioritization.
Conclusively, using three different validation sets we show that even for such a complex trait as schizophrenia GPrior can recover novel biologically relevant genes using only previously published data. Furthermore, using GPrior we can confirm significant overlap between gene sets acting through common or rare variants on schizophrenia risks.
MAGMA comparison
We compared GPrior with a commonly used method that takes GWAS summary statistics as input and attempts to pinpoint likely disease genes—MAGMA [26]. It computes gene-based p value (mean association of SNPs in the gene, corrected for LD). We ran MAGMA with default parameters and compared performance quality with GPrior. As an output MAGMA returns a list of genes and corresponding p-values, which we used to sort the list for prioritization purposes.
One of the challenges for a non-biased comparison was the relatively small number of gene candidates in output from MAGMA. Therefore, we took the same number of top genes from GPrior results to compare an equal number of gene predictions.
GPrior demonstrated enrichment of top ranked predictions for validation sets for all phenotypes–EA (p value = 9 × 10−3), schizophrenia (p value = 7 × 10−4), CAD (p value = 3 × 10−3) and IBD (p value = 0.019). MAGMA produced significantly enriched predictions only for CAD (p value = 9 × 10−3) (Supplementary Fig. S11).
Conclusively, GPrior demonstrated the best performance out of all evaluated approaches for gene prioritization in multiple settings and for various phenotypes.
Discussion
A large number of GWAS studies performed to date provide an invaluable source of information for generating biological hypotheses for disease causes. The majority of these studies have greatly benefitted from fine-mapping that implicated a limited number of gene candidates. However, for highly polygenic phenotypes like schizophrenia, known genes represent only a tiny segment of the disease biology. Current methodologies are unable to precisely separate true effector transcripts from nearby non-causal genes within associated GWAS loci.
The challenge of mapping “variants to function” can in principle greatly benefit from machine learning approaches—particularly for those phenotypes for which a strictly genetic fine-mapping approach has had limited success in conclusively identifying risk genes. As we illustrate, conventional positive-negative machine learning approaches require a substantial fraction of already known disease genes to achieve sufficient prioritization quality for novel candidates. In addition, it is nearly impossible at this point to confidently state that a gene is not involved in a disease, therefore, directly assuming “negative” examples for training is fated to include false negatives in a training set, further reducing prediction quality.
Instead, we provide a tool that uses positive-unlabeled learning and requires only confidence in selecting positive instances for training. Such genes are relatively easy to identify based on association significance, previously reported functional studies, etc. Importantly, PU-learning performs well even when the training set is quite small.
An additional challenge for a single-method-based solution is presented by phenotype complexity. Phenotypes may present significantly different genetic architectures or impose certain limitations on the set of available data sources; therefore, it is unlikely that a single technique will be suitable for gene prioritization in all of them. We provide a software package for gene prioritization – GPrior that takes advantage of the ensemble of PU-learning techniques. Such an approach overcomes the unresolved challenges of PN-learning and issues arising from phenotype complexity. In GPrior, two key steps of the model training: PU-classifiers training and selection of optimal classifiers composition are performed using two independent gene sets. The two-step strategy ensures independent quality assessment for all classifiers and unbiased selection of the optimal prioritization method, as well as delivering optimal prioritization results for the specific phenotype.
Several limitations of the method should be mentioned. First, there are a great number of understudied phenotypes, for which assembly of a reliable set of “gold-standard” disease genes that could be used for training still imposes a challenge. Therefore, using several gene sets (TS and AES) for training a model is nearly impossible due to the lack of the known genes. For this reason, we have an option to run GPrior using a single True set, without finding the optimal composition of predictions based on the algorithm evaluation set, and to obtain the prioritizations from all five algorithms for further manual analysis.
Further, the lack of a complete set of causal and non-causal genes for a trait significantly limits the validation procedure of gene prioritization results. We lack reliable benchmark data for gene prioritization, thus limiting the scope of validation experiments and introducing undesirable contamination in most of the commonly used quality scores. Using a clinically diverse set of well-studied phenotypes, we demonstrate the broad utility of the approach and validate the disease-specific nature of the resulting predictions in case studies (introducing noise into the training data and switching validation sets between genetically distant phenotypes).
Also, one of the limiting steps in our GWAS processing scheme was naïve and inclusive selection of gene candidates from each locus. More sophisticated preprocessing of the raw GWAS summary statistics with methods such as SuSie or FINEMAP to improve variant-to-gene mapping could significantly aid variant-level to gene-level features transformation.
Finally, as with any data-driven classifier, the predictions would be naturally biased by the features that were selected for input. For example, functional interactions derived from the Reactome are naturally biased towards more studied genes. Furthermore, we have not selected features to be specific to each phenotype. We used a relatively conservative set of features for gene annotations, which could be significantly expanded or replaced with more relevant phenotype-specific annotations, such as – single-cell expression data, specific protein–protein interactions, gene conservation metrics (pLI, LOEUF), and others. From the analysis of features importance we concluded that SNP-level features contributed the most in the resulting GPrior gene prioritization (e.g. VEP, Fantom5, DHS), but at the same time, there is a strong trait specificity in the importance of gene expression features. For example, IBD features importance analysis showed that gene expression in the Colon and Esophagus appears to be important for prioritizing true disease genes in comparison with other tissues (Supplementary Tables S15-S18). Therefore, users can expect to see higher performance in case of thorough feature selection. Additionally, GPrior can be straightforwardly integrated with conventional fine-mapping tools or other prioritization methods. Altogether, we certainly recommend using phenotype-specific annotations along with manual feature curation to achieve the best possible result.
Conclusively, GPrior fills an important and currently underdeveloped niche of methods for GWAS data post-processing, significantly improving the ability to pinpoint disease genes compared to existing solutions.
Code availability
References
Ding K, Kullo IJ. Methods for the selection of tagging SNPs: A comparison of tagging efficiency and performance. Eur J Hum Genet. 2007;15:228–36.
Foulkes AS. Applied statistical genetics with R. New York: Springer New York; 2009. https://doi.org/10.1007/978-0-387-89554-3.
Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Mol Genet. 2015;24:R111–R119.
Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–90.
Benner C, Spencer CCA, Havulinna AS, Salomaa V, Ripatti S, Pirinen M. FINEMAP: Efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–501.
Kichaev G, Yang WY, Lindstrom S, Hormozdiari F, Eskin E, Price AL, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:1004722.
Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–73.
Wang G, Sarkar A, Carbonetto P, Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J R Stat Soc Ser B. 2020;82:1273–300.
Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273.
Peat G, Jones W, Nuhn M, Marugán JC, Newell W, Dunham I, et al. The open targets post-GWAS analysis pipeline. Bioinformatics. 2020;36:2936–7.
Erratum: Genetic effects on gene expression across human tissues (Nature (2017) 550 (204-13). Nature. 2018;553:530.
Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7.
Bromberg Y. Chapter 15: disease gene prioritization. PLoS Comput Biol. 2013;9:e1002902.
Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinform. 2005; 6. https://doi.org/10.1186/1471-2105-6-55.
Xu J, Li Y. Discovering disease-genes by topological features in human protein–protein interaction network. Bioinformatics. 2006;22:2800–5.
Smalter A, Seak FL, Chen XW Human disease-gene classification with integrative sequence-based and topological features of protein-protein interaction networks. In: Proceedings of the 2007 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2007. 2007, pp. 209–14.
Isakov O, Dotan I, Ben-Shachar S. Machine learning–based gene prioritization identifies novel candidate risk genes for inflammatory bowel disease. Inflamm Bowel Dis. 2017;23:1516–23.
Denis F PAC learning from positive statistical queries*. In: Lecture notes in computer science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer Verlag; 1998. pp. 112–26.
Sriphaew K, Takamura H, Okumura M. Cool blog classification from positive and unlabeled examples. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Berlin, Heidelberg: Springer; 2009. pp. 62–73.
Bekker J, Davis J. Learning from positive and unlabeled data: a survey. Mach Learn. 2020;109:719–60.
Mordelet F, Vert JP. A bagging SVM to learn from positive and unlabeled examples. Pattern Recognit Lett. 2014;37:201–9.
Yang P, Li X, Chua H-N, Kwoh C-K, Ng S-K. Ensemble positive unlabeled learning for disease gene identification. PLoS One. 2014;9:e97079.
Scott C, Blanchard G. Novelty detection: unlabeled data definitely help. In: van Dyk D, Welling M (eds). Proceedings of the Twelth International Conference on Artificial Intelligence and Statistics. PMLR: Hilton Clearwater Beach Resort, Clearwater Beach, Florida USA, 2009, pp. 464–71.
Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009; 37. https://doi.org/10.1093/nar/gkp427.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput Biol. 2015;11:e1004219.
Lehne B, Lewis CM, Schlitt T. From SNPs to genes: disease association at the gene level. PLoS One. 2011;6:e20133.
Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24.
Ala U, Piro RM, Grassi E, Damasco C, Silengo L, Oti M, et al. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLoS Comput Biol. 2008;4:e1000043.
Fine RS, Pers TH, Amariuta T, Raychaudhuri S, Hirschhorn JN. Benchmarker: an unbiased, association-data-driven strategy to evaluate gene prioritization algorithms. Am J Hum Genet. 2019;104:1025–39.
Lee WS, Liu B. Learning with positive and unlabeled examples using weighted logistic regression. In: Proceedings of the Twentieth International Conference on Machine Learning (ICML). 2003. p. 2003.
Claesen M, De Smet F, Suykens JAK, De Moor B. A robust ensemble approach to learn from positive and unlabeled data using SVM base models. Neurocomputing. 2015;160:73–84.
Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86.
Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–8.
Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578:527–39.
Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73.
Momozawa Y, Dmitrieva J, Théâtre E, Deffontaine V, Rahmouni S, Charloteaux B, et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat Commun. 2018;9:1–18.
Liu JZ, Van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86.
Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–21.
Kaplanis J, Samocha KE, Wiel L, Zhang Z, Arvai KJ, Eberhardt RY, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–62.
Van Der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–43.
Khera AV, Kathiresan S. Genetics of coronary artery disease: Discovery, biology and clinical translation. Nat Rev Genet 2017;18:331–44.
Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9.
Singh T, Poterba T, Curtis D, Akil H, Eissa M Al, Barchas JD et al. Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. medRxiv. 2020; 2020.09.18.20192815.
Tang J, Chen X, Xu X, Wu R, Zhao J, Hu Z, et al. Significant linkage and association between a functional (GT)n polymorphism in promoter of the N-methyl-d-aspartate receptor subunit gene (GRIN2A) and schizophrenia. Neurosci Lett. 2006;409:80–2.
Koide T, Banno M, Aleksic B, Yamashita S, Kikuchi T, Kohmura K, et al. Correction: Common Variants in MAGI2 Gene Are Associated with Increased Risk for Cognitive Impairment in Schizophrenic Patients. PLoS One. 2012; 7. https://doi.org/10.1371/annotation/47ca9c23-9fdd-47f6-9d36-db0a31769f22.
Pinacho R, Saia G, Meana JJ, Gill G, Ramos B. Transcription factor SP4 phosphorylation is altered in the postmortem cerebellum of bipolar disorder and schizophrenia subjects. Eur Neuropsychopharmacol. 2015;25:1650–60.
Ripke S, Walters JTR, O’Donovan MC. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv. 2020; 2020.09.12.20192922.
Acknowledgements
The authors would like to thank Dr. Alexey Sergushichev (ITMO University) and Dr. Maxim Artyomov (Washington University in St. Louis) for helpful discussions.
Funding
N.K. was supported by the grant of the Ministry of Science and Higher Education of the Russian Federation (Agreement No. 075-15-2020-901).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study did not require ethical approval as no human subject data was involved.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kolosov, N., Daly, M.J. & Artomov, M. Prioritization of disease genes from GWAS using ensemble-based positive-unlabeled learning. Eur J Hum Genet 29, 1527–1535 (2021). https://doi.org/10.1038/s41431-021-00930-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-021-00930-w
This article is cited by
-
Tissue specific tumor-gene link prediction through sampling based GNN using a heterogeneous network
Medical & Biological Engineering & Computing (2024)
-
E-GWAS: an ensemble-like GWAS strategy that provides effective control over false positive rates without decreasing true positives
Genetics Selection Evolution (2023)
-
A novel candidate disease gene prioritization method using deep graph convolutional networks and semi-supervised learning
BMC Bioinformatics (2022)
-
Exploring Artificial Intelligence in Drug Discovery: A Comprehensive Review
Archives of Computational Methods in Engineering (2022)