Abstract
BRAF and MLH1 promoter methylation testings have been proven effective prescreens for Lynch Syndrome. We aimed to compare different screening strategies for Lynch Syndrome in patients with MLH1(−) CRC. Patients with MLH1(−) CRC who had been tested for BRAF mutation and germline variants of DNA mismatch repair genes were included. We compared the sensitivities and specificities for identifying Lynch Syndrome and the cost-effectiveness of four screening approaches that used the following tests as prescreens: BRAF testing, MLH1 methylation testing, MLH1 methylation & BRAF testing, and MLH1 methylation testing & Revised Bethesda Criteria. Of 109 patients included, 23 (21.1%) were Lynch Syndrome patients. BRAF mutation and MLH1 methylation occurred in 6 (5.5%) and 40 (36.7%) patients, respectively. The sensitivity for identifying Lynch syndrome of BRAF testing was 100%, but the specificity was only 7%. MLH1 methylation testing had a lower sensitivity than BRAF testing (97.5% vs 100%), but had a markedly higher specificity (45.3% vs 7%). The combination of the two testings had a slightly higher specificity than MLH1 methylation testing alone (47.7% vs 45.3%). The MLH1 methylation testing approach had a 10% lower cost of identifying MLH1(−) Lynch syndrome carriers per case than universal genetic testing, but it missed 4.5% of patients. BRAF and MLH1 promoter methylation testings as prescreens for Lynch syndrome are less effective in Chinese patients with MLH1(−) CRC than in their Western counterparts. Universal genetic testing could be considered an up-front option for this population.
Similar content being viewed by others
Introduction
Lynch Syndrome is the most common hereditary colorectal cancer (CRC) syndrome. Although it accounts for only 2%–3% of all CRCs, more than 50% of its carriers develop CRC or other tumors during their lifetimes [1]. Screening for Lynch Syndrome among CRC patients is important for early diagnosis and disease management. Indeed, a universal screening approach has been implemented for newly diagnosed CRC patients, which involves the analysis of miscrosatellite instability (MSI) or of the expression of DNA mismatch repair (MMR) proteins (MLH1, MSH2, MSH6, and PMS2) by immunohistochemistry [2]. About 15% of CRCs show high microsatellite instability (MSI-H) or do not express one or more of the four MMR proteins, leading to a status called MMR deficiency (dMMR).
Of the four MMR deficiencies, MLH1 deficiency represents a special subtype. It is the most common type of dMMR, accounting for more than 70% of the total [3, 4]. In addition to germline variants in MMR genes, MLH1 deficiency can also result from double somatic mutations in MLH1 and, more often, from the methylation of MLH1 promoter [5, 6]. MLH1 methylation is responsible for most of MLH1 deficiency observed in sporadic CRC cases; it can be used as a marker to exclude this population from unnecessary genetic testing. In addition, MLH1 promoter methylation is strongly associated with BRAF mutation, which makes BRAF testing an alternative tool for screening out Lynch Syndrome [7,8,9,10,11].
However, the value of these tests can be affected by the genetic disparity of CRCs between populations. For example, BRAF V600E mutation occurs in 15%-20% of Western CRC patients, but in less than 5% of Chinese CRC patients [12,13,14]. This low frequency is likely to reduce the value of BRAF testing for Chinese CRC patients, but there is no study evaluating these screening methods or comparing them with universal genetic testing among this population. Our study aims to compare different screening strategies for Lynch Syndrome in Chinese patients with MLH1(−) CRC.
Methods
Data collection
We reviewed a consecutive series of 3330 patients who were diagnosed as CRC in Sun Yat-sen University Cancer Center between November 1, 2011 and December 31, 2015. Immunotaining for MLH1, MSH2, MSH6, and PMS2 was performed for 3250 patients, 330 of whom were MMR deficient. Of the 330 patients, 170 were MLH1-deficient. Of them, 136 had undergone genetic testing with a panel that included MLH1, MSH2, MSH6, PMS2, and EPCAM in our previous study [15]. 109/136 patients had viable tumor tissue, and were included in the study.
Clinical data were collected from hospital records, including sex, age, personal history of cancer, family history of cancer, and pathology of tumors. This retrospective study was approved by the institutional review board of Sun Yat-sen University Cancer Center, and written informed consent was obtained from all patients.
Testing protocols
BRAF V600E testing
Somatic testing for BRAF mutation had been carried out for 165/170 patients with MLH1 deficiency by using formalin-fixed paraffin-embedded (FFPE) tissue blocks, according to the screening algorithm in our previous study [15]. All patients included in this study (n = 109) had undergone the test. Briefly, the BRAF mutation within exon 15 was tested using fluorescent real-time polymerase chain reaction (PCR). Genomic DNA was amplified in a 24-μl PCR reaction with 7500 real-time fluorescence quantitative PCR system (Applied Biosystems, Foster City, CA). Mutations were confirmed with independent duplicate analyses.
MLH1 promoter methylation testing
MLH1 promoter methylation testing was carried out by pyrosequencing for all 109 patients. Tumor DNA was extracted by QIAamp DNA FFPE Tissue Kit (QIAGEN, US), and then converted into bisulfite using EpiTect Bisulfite Kits (QIAGEN, US). After tumor DNA was purified, MLH1 promoter methylation testing was carried out with validated PCR primers specific to methylated sequences: MLH1-F:GTATTTTTGTTTTTATTGGTTGGATA;MLH1-R: CCAATCAAATTTCTCAACTCTATA. Fifty-cycle PCR was performed as follows: denaturation 95 °C, 15 s; annealing 54 °C, 20 s; extension 72 °C, 30 s. After DNA amplification, the DNA sequence was analyzed by PyroMark Q96 ID. The sequencing section contains 5 CpG: AGAGCGGACAGCGATCTCTAACGCGCAA-GCGCA.
Genetic testing of MMR genes
Genetic testing for hereditary colorectal syndromes had been undertaken for 109 patients, as previously described [15]. In brief, DNAs extracted from peripheral blood samples were used to construct libraries, which were then sequenced by a multigene panel covering MLH1, MSH2, MSH6, PMS2, and EPCAM. Lynch Syndrome was diagnosed when pathogenic or likely pathogenic variants in one of the five genes were identified.
Screening approaches and cost-effectiveness analyses
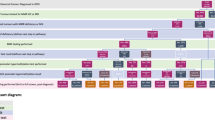
As shown in Fig. 1, our study used four approaches to screen for Lynch Syndrome. They were as follows:
Approach 1: BRAF mutation-germline testing. BRAF testing was used as a primary screening tool. Patients with BRAF mutation tumors were considered sporadic and were excluded, while patients without BRAF mutation proceeded with genetic testing for MMR genes.
Approach 2: MLH1 methylation-germline testing. Patients were screened for MLH1 promoter methylation before germline sequencing. Patients with methylated tumors were considered sporadic and excluded, while those without methylation were tested for germline variants.
Approach 3: BRAF mutation & MLH1 methylation-germline testing. Patients were pre-screened for BRAF mutation, and those without mutation were tested for MLH1 methylation. Germline sequencing was reserved for patients negative for both tests.
Approach 4: Bethesda Criteria & MLH1 methylation-germline testing. Patients were prescreened with the Revised Bethesda Criteria. Patients who did not met the criteria underwent MLH1 methylation testing. Patients who met the criteria and patients without MLH1 methylation were recommended for genetic testing.
We then performed cost-effectiveness analyses for the four approaches and compared their costs of identifying MLH1(−) Lynch syndrome patients per case. The prices of BRAF testing and MLH1 methylation testing were 472 RMB and 708 RMB per case, respectively, and the price of genetic testing with a 14-gene panel was 3220 RMB.
Statistics
Associations between categorical variables were assessed by chi-square test, Fisher’s exact test, or Wilcoxon’s rank-sum test, as appropriate. We compared the sensitivities and specificities for identifying Lynch Syndrome, positive predictive values (PPVs), and negative predictive values (NPVs) of the four screening approaches. All P values were two-sided, and P < 0.05 was considered statistically significant. All of the analyses were performed in R (version 3.5.1).
Results
Baseline information
Table 1 shows the baseline information. Of the 109 MLH1-deficient patients, 49 (45.0%) were female; the median age at diagnosis was 54.0 years. BRAF mutation and MLH1 methylation were observed in 6 (5.5%) and 40 (36.7%) patients, respectively. Germline variants in MMR-related genes were found in 49 (45.0%) patients; among those, 11 were pathogenic, 12 were likely pathogenic, and 26 were variants of unknown significance (VUS). All pathogenic and likely pathogenic variants occurred in MLH1 (see Supplementary Table 1). In total, 23 (21.1%) patients were diagnosed with Lynch Syndrome, and the other 86 (78.9%) with sporadic CRC.
Comparison of characteristics between Lynch and sporadic CRCs
Table 2 shows the comparisons between Lynch and sporadic CRC. Compared with sporadic CRC, Lynch Syndrome related CRC was associated with a younger age (46.8 vs 55.8 years), fewer mucinous carcinomas (17.4% vs 44.2%), and higher prevalence of cancer history among first-degree relatives (65.2% vs 23.3%). BRAF mutation was observed only in sporadic CRC. Most of MLH1 methylation occurred in sporadic CRC, with one exception (39 vs 1, P < 0.001). This patient was a 47-year-old male, with personal and family histories of CRC, fulfilling revised Bethesda Criteria; he harbored pathogenic variant in MLH1 (Exon 8, c.677G>A). There were no differences in terms of sex, sideness of primary tumor, and history of polyps.
Performance of different screening approaches
BRAF mutation-genetic testing
BRAF mutation was identified in six sporadic CRC patients. The sensitivity for identifying Lynch Syndrome of this approach was 100%, but the specificity was only 7%. The positive predictive value (PPV) and negative predictive value (NPV) of this test were 22.3% and 100%, respectively (Table 3).
MLH1 methylation-genetic testing
MLH1 promoter methylation was identified in 40 (36.7%) patients, with 39 (97.5%) being sporadic. The sensitivity and specificity for identifying Lynch Syndrome of this approach were 95.7% and 45.3%, respectively (Table 3). This approach missed 4.3% of Lynch Syndrome carriers. The PPV of this test was 31.9%, slightly higher than that of the BRAF testing approach. MLH1 methylation and germline variants together account for 56.9% of MLH1 deficiency, while the other 43.1% remained unexplained.
Combination of MLH1 promoter methylation testing with BRAF testing and with Bethesda criteria
Five (4.6%) patients with MLH1 methylation were BRAF mutant-type; 68 (62.4%) patients without MLH1 methylation were BRAF wild-type. The concordance rate of the two tests was 67.0%. When BRAF and MLH1 methylation tests were combined, the addition of BRAF testing did not improved the sensitivity for identifying Lynch Syndrome of MLH1 methylation testing, with 4.3% of Lynch Syndrome carriers remaining missed. When the Revised Bethesda Criteria and MLH1 methylation testing were combined, the sensitivity increased to 100%, with no Lynch Syndrome patient being missed, but the PPV declined from 31.9% to 27.4%, suggesting that an additional 4.5% patients would be referred for unnecessary genetic testing.
Cost-effectiveness analyses
Cost-effectiveness analyses are showed in Table 4. Among the four pre-screening approaches, MLH1 methylation testing had the lowest costs of identifying MLH1(−) Lynch syndrome per case. The costs of this approach were lower than those of universal genetic testing (13,607 RMB vs 15,260 RMB), but its omission rate was higher (4.3% vs 0).
Discussion
Our study found that BRAF mutation was rare among MLH1 deficient CRCs in the Chinese population. Germline variants in MMR genes and MLH1 promoter methylation together accounted for only 56.9% of MLH1 deficiency. BRAF testing and MLH1 promoter methylation testing as prescreens to exclude Lynch Syndrome were less effective in Chinese CRC patients than in Western CRC patients.
MLH1 methylation has been proposed as a screening marker for excluding patients from germline testing, because it occurs mostly in non-Lynch MLH1(−) CRCs. Tomer et al. found that MLH1 methylation existed in 85.5% of MLH1(−) CRCs, and its detection helped to reduce the rate of referral to genetic testing from 31% to 13.5% [16]. In our study, however, MLH1 methylation was identified in less than 40% of the MLH1(−) CRCs, leading to a significant reduction in its utility as an exclusion criterion for Lynch syndrome.
MLH1 methylation can coexist with Lynch Syndrome. A previous study found that MLH1 methylation occurred in 15% of Lynch Syndrome carriers, who would be missed when using MLH1 methylation as an exclusion criterion [17]. In our study, 39/40 (97.5%) patients with MLH1 methylation were sporadic; only one (2.5%) patient with Lynch syndrome was missed. It appears that Chinese Lynch Syndrome carriers are less likely to harbor MLH1 methylation compared with their Western counterparts, which explains the high sensitivity for identifying Lynch syndrome of this approach in our study.
A simpler screening method is to detect BRAF mutation. BRAF mutation is highly concordant with MLH1 methylation, and can also be used as an exclusion criterion. Bessa et al. found that BRAF testing could improve the cost-effectiveness of genetic testing for Lynch Syndrome [18]. Moreover, in a Spanish study, BRAF testing was more sensitive than MLH1 methylation testing for ruling out Lynch Syndrome [17]. However, in some studies, BRAF testing was less effective than MLH1 methylation testing, identifying only 20–40% of sporadic CRCs [18, 19]. In Chinese CRCs, this number may further shrink because BRAF mutation occurs in less than 5% of Chinese CRCs [13, 20]. In our study, BRAF mutation was observed in 5.5% of the patients and in 15% (6/40) of the MLH1-methylated cases; therefore BRAF testing spared only a small number of patients from unnecessary genetic testing. The results are consistent with a previous Chinese study [21], in which BRAF mutation was found in only 15% of Chinese patients with MLH1-/PMS2- CRC, significantly lower than that in their Western counterparts (63%) [22]. Therefore, BRAF mutation testing had little to offer as a prescreen to rule out sporadic cases in Chinese patients with MLH1(−) CRC.
A previous study found that compared with MLH1 methylation testing alone, the combination of BRAF testing and MLH1 methylation testing could slightly reduce the rate of referral to genetic counseling [16]. In our study, although the addition of BRAF testing slightly increased the specificity of MLH1 methylation testing (from 45.3% to 47.7%), it failed to improve the sensitivity, with one Lynch Syndrome carrier remaining missed. Therefore, the performance of this combination is similar to that of MLH1 methylation testing alone.
To improve the performance of MLH1 methylation testing, we introduced the Revised Bethesda Criteria as a first-step screening to select eligible patients. As expected, the criteria helped to include the Lynch Syndrome case missed by MLH1 methylation testing, increasing its sensitivity to 100%. However, the criteria also included a considerable number of low-risk cases for genetic testing, leading to a drop in PPV from 31.9% to 27.4%. Although this combination increased the sensitivity for identifying Lynch Syndrome, it also increased the costs.
Genetic testing is the gold standard for diagnosing Lynch Syndrome, but it is used mainly as a diagnostic tool rather than a screening one because of its high costs. However, as the technology matures and the price goes down, it can also be used in screening programs. In 2012, Wang et al. found that genetic testing for first-degree relatives of Lynch Syndrome patients, who were at high risk of CRC, was a cost-effective strategy in Singapore [23]. Given the poor performance of BRAF and MLH1 promoter methylation testing in excluding low-risk CRCs, it is reasonable and practical to carry out universal genetic testing for MLH1(−) CRCs among the Chinese population. Although in cost-effectiveness analysis, the MLH1 methylation testing screening approach had lower costs for the identification of MLH1(−) Lynch syndrome per case than universal genetic sequencing, it should be noted that the costs of genetic testing in our study cover 14 genes related to hereditary CRC syndromes. When this panel narrows down to five genes (MLH1, MSH2, MSH6, PMS2, and EPCAM), or even two (MLH1 and PMS2), the gap between these two approaches may likely close.
Our study has some limitations. First, as a single-center study, it is subject to selection bias. Second, the sample of this study is relatively small, precluding it from drawing definitive conclusions. Third, family history of cancer is unavailable in some patients, which may weaken the performance of the Revised Bethesda Criteria. Last, somatic sequencing and germline MLH1 methylation testing were not performed in this study, and therefore we were not able to assess other causes for MLH1 deficiency.
Conclusion
Taken together, our study demonstrates that BRAF and MLH1 promoter methylation testing as prescreening approaches for Lynch Syndrome among patients with MLH1(−) CRC are less effective in the Chinese population than in the Western population. MLH1 methylation testing as a prescreen helped to reduce the costs of identifying MLH1(−) Lynch syndrome per case by about 10% percent compared with universal genetic testing, but it ran the risk of omitting 4.3% of Lynch syndrome carriers. Universal genetic testing could be considered an up-front option for this population.
References
Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, Longy M, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305:2304–10.
Moreira L, Balaguer F, Lindor N, de la Chapelle A, Hampel H, Aaltonen LA, et al. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308:1555–65.
Pérez-Carbonell L, Ruiz-Ponte C, Guarinos C, Alenda C, Payá A, Brea A, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012;61:865–72.
Ward RL, Hicks S, Hawkins NJ. Population-based molecular screening for Lynch syndrome: Implications for personalized medicine. J Clin Oncol. 2013;31:2554–62.
Palomaki GE, McClain MR, Melillo S, Hampel HL, Thibodeau SN. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med. 2009;11:42–65.
Sourrouille I, Coulet F, Lefevre JH, Colas C, Eyries M, Svrcek M, et al. Somatic mosaicism and double somatic hits can lead to MSI colorectal tumors. Fam Cancer. 2013;12:27–33.
Giardiello FM, Allen JI, Axilbund JE, Boland CR, Burke CA, Burt RW, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol. 2014;109:1159–79.
Stoffel EM, Mangu PB, Gruber SB, Hamilton SR, Kalady MF, Lau MWY, et al. Hereditary colorectal cancer syndromes: American Society of Clinical Oncology Clinical Practice Guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology Clinical Practice Guidelines. J Clin Oncol. 2015;33:209–17.
Rubenstein JH, Enns R, Heidelbaugh J, Barkun A, Adams MA, Dorn SD, et al. American Gastroenterological Association Institute Guideline on the diagnosis and management of lynch syndrome. Gastroenterology. 2015;149:777–82.
Kidambi TD, Blanco A, Myers M, Conrad P, Loranger K, Terdiman JP. Selective versus universal screening for lynch syndrome: a six-year clinical experience. Dig Dis Sci. 2015;60:2463–9.
Toon CW, Walsh MD, Chou A, Capper D, Clarkson A, Sioson L, et al. BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol. 2013;37:1592–602.
Yoon HH, Shi Q, Alberts SR, Goldberg RM, Thibodeau SN, Sargent DJ et al. Racial Differences in BRAF/KRAS Mutation Rates and Survival in Stage III Colon Cancer Patients. J Natl Cancer Inst. 2015; 107.
Guo T-A, Wu Y-C, Tan C, Jin Y-T, Sheng W-Q, Cai S-J, et al. Clinicopathologic features and prognostic value of KRAS, NRAS and BRAF mutations and DNA mismatch repair status: a single-center retrospective study of 1,834 Chinese patients with Stage I-IV colorectal cancer. Int J Cancer. 2019;145:1625–34.
Vilkin A, Niv Y, Nagasaka T, Morgenstern S, Levi Z, Fireman Z, et al. Microsatellite instability, MLH1 promoter methylation, and BRAF mutation analysis in sporadic colorectal cancers of different ethnic groups in Israel. Cancer. 2009;115:760–9.
Jiang W, Cai M-Y, Li S-Y, Bei J-X, Wang F, Hampel H, et al. Universal screening for Lynch syndrome in a large consecutive cohort of Chinese colorectal cancer patients: high prevalence and unique molecular features: Universal screening for LS in Chinese CRC patients. Int J Cancer. 2019;144:2161–8.
Adar T, Rodgers LH, Shannon KM, Yoshida M, Ma T, Mattia A, et al. A tailored approach to BRAF and MLH1 methylation testing in a universal screening program for Lynch syndrome. Mod Pathol. 2017;30:440–7.
Moreira L, Muñoz J, Cuatrecasas M, Quintanilla I, Leoz ML, Carballal S, et al. Prevalence of somatic mutl homolog 1 promoter hypermethylation in Lynch syndrome colorectal cancer: MLH1 Methylation in Lynch Syndrome CRC. Cancer. 2015;121:1395–404.
Bessa X, Ballesté B, Andreu M, Castells A, Bellosillo B, Balaguer F, et al. A prospective, multicenter, population-based study of BRAF mutational analysis for Lynch syndrome screening. Clin Gastroenterol Hepatol. 2008;6:206–14.
Pérez-Carbonell L, Alenda C, Payá A, Castillejo A, Barberá VM, Guillén C, et al. Methylation analysis of MLH1 improves the selection of patients for genetic testing in Lynch syndrome. J Mol Diagn. 2010;12:498–504.
Chen J, Guo F, Shi X, Zhang L, Zhang A, Jin H, et al. BRAF V600E mutation and KRAS codon 13 mutations predict poor survival in Chinese colorectal cancer patients. BMC Cancer. 2014;14:802.
Ye J-X, Liu Y, Qin Y, Zhong H-H, Yi W-N, Shi X-Y. KRAS and BRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World J Gastroenterol. 2015;21:1595–605.
Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49:151–7.
Wang VW, Koh PK, Chow WL, Lim JFY. Predictive genetic testing of first degree relatives of mutation carriers is a cost-effective strategy in preventing hereditary non-polyposis colorectal cancer in Singapore. Fam Cancer. 2012;11:279–89.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (No. 81871971 to PD), the Science and Technology Planning Project of Guangzhou City of China (No. 201803010117 to PD), and Science and Technology Program Project of Guangzhou (No. 201802020030 to LM). We also thank the Research Data Deposit public platform (www.researchdata.org.cn) for validating the authenticity of the data of this article (the approval number RDDA2020001429).
Author information
Authors and Affiliations
Contributions
Conceptualization: P-RD and WJ; Methodology: JL, BX and LM; Formal analysis and investigation: JL, EX; Resources: EX, LK, QS, DL, WL, ZH and JT; Writing—original draft preparation: BX and JL; Writing—review and editing: BX and PD; Supervision: WJ and PD; Project administration: ZP and PD; Funding acquisition: ZP and PD.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Xiao, B., Luo, J., Xie, E. et al. Comparisons of screening strategies for identifying Lynch syndrome among patients with MLH1-deficient colorectal cancer. Eur J Hum Genet 28, 1555–1562 (2020). https://doi.org/10.1038/s41431-020-0687-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-0687-8