Abstract
The X-linked recessive degenerative disease dystrophinopathy results from variants in the DMD gene. Given the large size and complexity of the DMD gene, molecular diagnosis for all dystrophinopathies remains challenging. Here we identified two cryptic exon retention variants caused by intronic single nucleotide variants in dystrophinopathy patients using combined RNA- and DNA-based methods. As one variant was previously unreported, we explored its likely pathogenic mechanism, via bioinformatic prediction for in silico verification of splicing. Then we constructed a minigene system harboring the variant and used morpholino modified antisense oligonucleotides (ASOs) to induce cryptic exon skipping. ASOs treatment corrected the mis-splicing in the mutant minigene system. Our study defines a novel intronic variant that can cause dystrophinopathy, and illustrates a strategy to overcome the aberrant splicing.
Similar content being viewed by others
Introduction
Dystrophinopathy is a type of X-linked recessive genetic disease that includes the more severe Duchenne muscular dystrophy (DMD; OMIM 310200), the less severe Becker muscular dystrophy (BMD; OMIM 300376), and the rare X-linked dilated cardiomyopathy (XLCM; OMIM 302045). The DMD gene is the largest known human gene to date, spanning ~2.2 M bases on chromosome Xp21.2. However, one of the full-length isoforms, Dp427m, is encoded by a nearly 14 kb transcript [1]. Consequently, >99% of the gene sequence is composed of noncoding sequences.
As we know, up to 70% variants in DMD are large deletion/duplications, 25–30% are small variants [2]. So for screening the DMD gene variants, the most common strategy is focused on coding regions, such as multiplex ligation-dependent probe amplification (MLPA) [3] and whole-exome sequencing (WES) [4], leaving deep intronic variants undetected. Pre-mRNA splicing is critical in gene expression process. It involves interactions between trans-acting factors and cis-regulatory elements. Trans-acting factors are splicing factor proteins and regulate RNA splicing by binding directly or indirectly to the cis-regulatory elements. Cis-regulatory elements are pre-mRNA sequences also known as splicing regulatory elements (SREs), such as exonic splicing enhancers (ESEs) that promote pre-mRNA splicing and exonic splicing silencers that repress the splicing [5]. Deep intronic single nucleotide substitutions sometimes generate new splicing sites or SREs, inducing the retention of a cryptic exon (CE), also mentioned as pseudoexon. The SRE can be targeted by an antisense oligonucleotide (ASO) to induced exon skipping. This strategy may provide possibility to prevent CE inclusion.
Here we report three patients from two families for whom typical methods (MLPA, WES) did not identify genetic call variants. After detailed clinical assessment, we undertook muscle biopsies and used immunohistochemical staining to ascertain appropriate diagnoses of dystrophinopathy. Hypothesizing that their causal variations were likely intronic, we examined transcripts for the presence of retained intronic sequences, also called CE, and then used targeted sequencing to identify the causal variant. As one variant is previously unreported, we explored its likely pathogenic mechanisms, initially via predictive bioinformatics and subsequently using a minigene system for in vitro determining splicing outcome of variant. Having confirmed it as a causal variant, we envisioned the use of morpholino modified ASOs to prevent the retention by CE skipping. This strategy worked, and the mis-splicing was corrected.
Subjects and methods
All three patients and potential carriers in their families were investigated. Written informed consent was obtained from each subject to use their DNA for molecular analysis of DMD gene and further research. For minor patients, parental consent for muscle biopsy and molecular analysis was obtained. The research was conducted according to the principles of the declaration of Helsinki. The study was approved by the ethics committee of First Affiliated Hospital of Fujian Medical University. The genotype data in this study are deposited on the Leiden Open Variation Database (LOVD V.3.0). The data are accessible with the individual ID#00266165, 00266125, and 00266152 on the website https://databases.lovd.nl/shared/individuals.
Clinical assessment
All patients were assessed according to the standard process suggested in the latest DMD Care Considerations Working Group guidelines [2]. The ambulatory ability was assessed by the North Star Ambulatory Assessment (NSAA) and 6-minute walking distance (6MWD) [6]. Blood creatine kinase (CK), blood/urinary myohemoglobin, electrocardiograph, echocardiogram, and pulmonary function were examined.
Muscle biopsy
Muscle specimens were collected from patients’ quadriceps femoris or deltoid after signing an informed consent form. Serial frozen sections were stained with hematoxylin and eosin (HE), modified Gomori trichrome, and nicotinamide adenine dinucleotide. The cross sections were stained by dystrophin antibodies including NCL-DYS1 (rod domain, clone Dy4/6D3), NCL-DYS2 (C terminus, clone Dy8/6C5,), and NCL-DYS3 (N terminus, clone Dy10/12B2) from Novocastra (Leica biosystems, Newcastle, United kingdom) according to the standard protocol.
Genetic analysis
Genomic DNA was extracted from peripheral leukocytes obtained from patients and their family members using DNeasy Blood and Tissue Kits (QIAGEN, Hilden, Germany). MLPA was conducted using a commercial kit SALSA MLPA Probemix P034 version B2 DMD-1 and P035 version B1 DMD-2 (MRC Holland, Amsterdam, The Netherlands). Exons and flanking splicing sites were amplified by primers according to Leiden Muscular Dystrophy data pages (http://www.dmd.nl/exonprim_seq.html) using ExTaq DNA polymerase (TaKaRa, Beijing, China), and analyzed by Sanger sequencing for exonic single nucleotide variants.
A sufficient quantity of archived muscle tissue was obtained from all patients to perform diagnostic RNA extraction and reverse transcription polymerase chain reaction (RT-PCR) based sequencing. Total RNA was extracted using Trizol reagent (Invitrogen, La Jolla, CA) according to the manufacturer’s recommended protocol, and complementary DNA (cDNA) was generated using a HiScript II Q RT SuperMix kit (Vazyme, Nanjing, China) according to the kit instructions. RT-PCR was performed using high-fidelity enzyme KOD-plus-Neo (TOYOBO, Osaka, Japan) with primers designed to amplify 22 overlapping fragments of the entire dystrophin mRNA (Table S1). Then, fragments were analyzed by Sanger sequencing.
In silico analysis of DMD sequences
The locations of ESEs in the sequences of patients were predicted using the program “ESEfinder” (version 3.0; http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) according to previous research [7]. This program can score the sequence as motifs thought to serve as binding sites for the specific serine/arginine (SR) splicing factors such as SF2/ASF, SC35, SRp40, and SRp55.
Splice-site score predictions for the CEs were performed using the Human Splicing Finder (HSF) Web interface (version 3.1; http://www.umd.be/HSF), which includes position weight matrices to calculate consensus values (CV) and an algorithm for the calculation of the MaxEnt scores [8].
DMD CE minigene construction
We constructed the normal and DMD CE minigene spanning GRCh38_chrX:32386273–32389738 including the last 64 bp of intron 31, exon 32, full-length intron 32, exon 33, and the first 37 bp of intron 33 (Fig. 3b). This sequence was amplified in three fragments from the human genome using high-fidelity enzyme KOD-plus-Neo (TOYOBO, Osaka, Japan) with the primers listed in Table S3. Then, we joined them together with a plasmid back-bone as described previously [9] into a minigene expressing vector using a ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China). The identity of minigene plasmids was verified by Sanger sequencing.
Cell culture and transfection
The HEK 293T cell line was cultured in Dulbecco’s modified Eagle’s Medium (Gibco, Grand Island, New York) consisting of 10% fetal bovine serum (Gibco, Grand Island, New York) and 1% penicillin/streptomycin (P/S) (Thermo Fisher Scientific, Waltham, Massachusetts). Cells were cultured at 37 °C in an incubator with a 5% CO2 atmosphere. Transfection was performed with Lipofectamine 3000 Reagent kit (Thermo Fisher Scientific, Waltham, Massachusetts) according to the manufacturer’s instructions. RNA extraction and RT-PCR were performed using methods described above. T-A cloning was employed to distinguish two fragments of alternative splicing using T-Vector pMD19 (TaKaRa, Beijing, China).
Cryptic exon skipping
According to the result of prediction by ESEfinder and HSF, we designed and synthesized three morpholino modified ASOs (Gene Tools, Philomath, Oregon) to suppress splicing sites and ESE, respectively (Table S4), for CE skipping. HEK 293T cells were transfected with morpholino modified ASOs 24 h after minigene plasmid transfection using Endo-Porter simple delivery reagent (Gene Tools, Philomath, Oregon), according to the manufacturer’s protocol. For RNA studies, cells were harvested using Trizol (Invitrogen, Dublin, Ireland) at the indicated time points. RT-PCR was conducted using methods described above. The RT-PCR products were separated on 1.5% agarose gels and visualized on a UV transilluminator (Tanon, Shanghai, China).
Results
Clinical manifestation
All three patients from two families presented clinical features consistent with a dystrophinopathy. Proband (II-1) in Family 1, a boy born in 2011, was referred to our hospital for gait abnormalities and elevated serum CK at the age of 6. He had jumping difficulties, and presented hypertrophic calves with Gowers’ sign, yet he could climb stairs without assistance. His 6MWD was 416 meters, and his score on the NSAA was 29. His intellectual development is normal. There were no similar patients in his family (Fig. 1a).
a–b Pedigree chart of dystrophinpathy families 1(A) and 2(B) with pseudoexon retention. Filled black squares indicate unambiguously affected individuals; hemi-black circles indicate female carriers; arrows indicate the probands. c Muscle tissue staining for II-1 in family 1, hematoxylin and eosin staining (HE) revealed mild fibrosis, degenerating, and regenerating. Immunohistochemical analysis (IHC) shows diffuse reduction of staining at the muscle membrane with DYS-2 and DYS-3, but had an almost normal staining pattern for an antibody directed to DYS-1. d Muscle tissue staining for IV-1 in family 2, HE revealed a typical dystrophic pattern including areas of fibrosis, sever infiltration of inflammatory cells, and obvious degenerating and regenerating. The IHC pattern showed no signal for dystrophin with DYS-1, DYS-2, and DYS-3.
Proband (IV-1) in family 2, a male child born in 2014, was diagnosed after initially being flagged based on his elevated CK level during a routine health examination. His exercise performance including running and jumping has been weaker than that of his peers since birth. He walked normally and did not need any assistance when climbing stairs, but showed mild exercise intolerance, proximal weakness, and pseudo-hypertrophic calves without Gowers’ sign. He has an X-linked family history of similar symptoms, and his uncle and two great-uncles suffered from proximal limb weakness and then lost their ambulation by their second decades, finally dying as teenagers (Fig. 1b). His elder brother (IV-3), born in 2010, showed typical clinical features for dystrophinopathy from age 4, including neck flexors and proximal limb weakness, pseudo-hypertrophic calves with Gowers’ sign, and frequent falls, but without mental retardation. Tip-toe walking started about age 7. He had been to many well-known hospitals in China, but no one could definitively diagnose the disease; no causative variant was detected even with DMD-MLPA and with next generation sequences of his whole exome. We ultimately ended up collected and analyzing DNA samples for his whole family.
Patients’ general information, movement presentation, and experimental test results are listed in Table 1. For all three patients, their pulmonary function, electrocardiographs, and echocardiograms appeared normal.
Immunohistochemistry analysis
Seeking confirmatory evidence for a dystrophinopathy diagnosis, we performed muscle biopsies for each of the three patients. Immunohistochemistry analysis of the muscle section from patient II-1 (family 1) showed a diffuse reduction of staining at the muscle membrane with standard antibodies directed to the N-terminal and C-terminal ends of the dystrophin protein, but had an almost normal staining pattern for an antibody directed to its rod domain (Fig. 1c), indicating a BMD diagnosis. This BMD diagnosis also fits with the clinical presentation of the patient. In contrast, the IHC pattern detected in the muscle tissue from patients IV-1 and IV-3 (family 2) lacked any signal for dystrophin (Fig. 1d), establishing a DMD diagnosis [10].
Identification of the causal variant
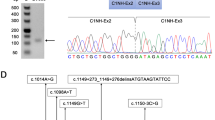
To test for the presence of variants in the DMD gene transcripts in these three patients, we extracted total RNA from muscle tissue biopsied from the patients and used the modified overlapping cDNA fragments amplified and separately sequenced strategy [11] used for studies of the DMD gene (Xp21.2 locus) to enable Sanger sequencing. We thusly identified retained sequences (Fig. 2a, b): a 58 BP sequence that BLAST analysis against the NCBI GRCh38 database revealed was precisely intercalated between exons 62 and 63, as well as two sequences that were each precisely intercalated between exons 32 and 33, one of 172-bp and one of 167-bp in size. The two retained sequences intercalated between exons 32 and 33 were both identified in tissue from a single patient in family 2. We also found that residual normal transcripts were present in patient II-1 (family 1) at a low level (Fig. 2a). Targeted primers (Table S2) based on the aforementioned BLAST analysis narrowed the likely variants to regions including intron 62 as well as intron 32. Sanger sequencing revealed that two variants can account for the observed intronic sequence retention (NG_012232.1(NM_004006.2): c.9225–285A>G, and c.4518 + 512T>A).
a–b RT-PCR of the muscle tissue derived mRNA, shows intronic sequence insertion between two exons. Two overlapping sequences can be recognized in family 2, following the sequence of exon 32; they can be distinguished as a 172 and 167-bp cryptic exon sequence starting with different splicing acceptor site. c–d Sanger sequencing confirmation in genomic DNA in patient and mother. See Table 2 for detail information of mutations, including genomic variant position, size and location of inserted fragments and protein change. e Splicing scheme. The picture shows normal splicing of exons and the expected splicing, which includes the pseudoexon between sequential exons. Dystrophin exons are represented as blue boxes; pseudoexons are represented as orange boxes. Arrows indicate positions of the mutations relative to the pseudoexons. The positions of the modified morpholino ASOs used for exon-skipping studies are indicated as 32N, 32E, and 32D.
Details of each variant including the position of each intronic fragment relative to the flanking exons are presented in Table 2. In each case, sequencing of the appropriate intron revealed the presence of point changes which apparently created novel splice sites. Specifically, II-1 in family 1 had a c. 9225–285A>G variant. IV-1 and IV-3 in family 2 both had a c. 4518 + 512T>A variant (Fig. 2c, d).
Confirmation by bioinformatics analysis and minigene system
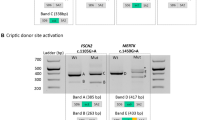
Bioinformatics analysis of the variant positioned in the CE was next conducted using both the HSF and ESEfinder. This c. 4518 + 512T>A variant creates a new 3′ss (HSF score varying from 54.09 to 83.04) that is used in the minority transcripts (CE of 167 bp) but also activates the use of an upstream preexisting 3′ss without changing its strength (score of 82.98 by HSF) leading to the inclusion of the 172-bp CE, likely due to the creation of an ASF/SF2-dependent ESE (Fig. 3a). ESEfinder predicted that the new variant may generate one new ESE motif (TACAGGA) whose score is calculated by two different matrices: 79.27 by standard SF2/ASF matrices (threshold 72.98) and 72.31 by IgM-BRCA1 matrices (threshold 70.51). It thus appears that the activation is likely due to a change in an SRE and not, or not only, the strengthening of a preexisting splice site.
a Bioinformatics analysis of reference and mutated sequences of the insert using “ESE finder3.0”. The program recognized two new high-score motifs (3.03 and 2.08, respectively) called SF2/ASF and SF2/ASF (IgM-BRAC1) indicated by arrows. The location of point mutation is framed on red. b Construction of minigene plasmid vector. The minigene compromised cytomegalovirus (CMV) promotor, 3.4 kb consecutive genomic DNA, elongation factor 1 alpha (EF1α) promotor, and green fluorescent protein (EGFP)sequence. c Agarose gel electrophoresis of RT-PCR products of minigene expression vector. d T-A clone of mutant minigene expression system. Green words indicate exon 32, red words indicate pseudoexon, and blue words indicate exon 33.
In the light of these predictions, we tested if the c. 4518 + 512 T>A variant does indeed promote retention of CE 32. We used the established minigene system [5] to generate CMV-target gene-EF1α-EGFR plasmids (Fig. 3b) bearing 3.4 kb wild type or c. 4518 + 512 T>A variant sequence for a gene region extending from exon 32 to exon 33 (GRCh38_chrX:32386273–32389738). We transfected HEK 293T cells with these plasmids, and 2 days later extracted RNA to assess the splicing pattern for the target region. As observed in the muscle tissue biopsied from the patients, our minigene assays revealed that the c. 4518 + 512 T>A variant promotes inclusion of this intronic region in dystrophin mRNA (Fig. 3c, d). Although there is some normal splicing occurring without treatment, it does not impact the ability of system to mimic the mis-splicing pattern.
ASOs mediated cryptic exon skipping
ASOs are short, single-stranded DNA molecules that interact with messenger RNA to modulate splicing. Splice modulating ASOs have been approved to treat spinal muscular atrophy and DMD. To investigate the feasibility using ASO as part of an antisense-mediated therapeutic CE-skipping strategy to ameliorate the observed pathogenic retention of the intronic sequence, we used the aforementioned minigene bearing HEK 293T cells and conducted assays with three morpholino modified ASOs designed to target CE: one was directed to the new ESEs and acceptor splicing site (32N), one was directed to the cluster of ESEs (32E) in the CE, and the last one was directed to the donor splicing site (32D).
The minigene transfected HEK 293T cells were treated with increasing amounts (from 5 to 20 nM) of three morpholino modified ASOs. We extracted total RNA 36 h later. RT-PCR analysis (Fig. 4) showed that ASO treatment increased the accumulation of normally spliced dystrophin mRNA; this increase was observed even at the lowest tested ASO concentration (5 nM), and exhibited a dose-dependent response. The 32E responded better than 32N and 32D for the treatment at 20 nM concentration, and cryptic splicing was almost corrected. 32N and 32D got almost top restoration at 10 nM concentration. There was no obvious further increase of normal splicing at 20 nM concentration.
Discussion
Molecular diagnosis for all dystrophinopathies can be quite challenging due to the huge size and complex structure of the DMD gene. It is reported that 2% [12] of variants cannot be detected by genomic variant analysis methods in dystrophinopathies. Without molecular diagnosis, prenatal diagnosis, preimplantation genetic diagnosis, and administration of gene therapies are unavailable. Thus, in these WES and MLPA negative cases, we eluded the typical genetic analysis strategy, diagnosed the disease by analyzing the muscle biopsy data, and got a positive finding in transcript. Finally, our study demonstrates the dystrophinopathy-causing variants, and even provides a strategy overcoming the aberrant splicing. In addition, from a basic science perspective, our characterization of the variant sheds new light on the mechanisms known for CE inclusion in disease.
Deep intronic variants leading to CE inclusion in the DMD gene were reported much less frequent compared with other types of DMD variants. In 1998, Ikezawa et al. first described a DMD patient who carried c. 3639 + 2240 A>G variant resulting in a 202 bp intronic sequence activation using muscle tissue RNA analysis [13]. Most variants were located at a splice site (±1–2 bp from the exon–intron boundary) [11, 14,15,16,17,18]. A single nucleotide substitution may complete the splice donor or acceptor of the potential CE, facilitating the activation of a probable cryptic splice site. In 2003, Tuffery-Giraud et al. characterized a BMD patient with mental retardation harboring a c. 9225–285 (+5 bp from the CE–intron boundary) A>G substitution, which resulted in the occurrence of a high-quality donor splice site for U1snRNA [15]. We identified the same variant in our BMD patient.
In 2011, Khelifi et al. reported two cases in which intronic fragment deletions could also result in the creation of novel splice sites, with CE incorporation into mRNA [19]. Then they constructed a minigene to confirm the pathology. In 2014, Trabelsi et al. discovered a pattern of CE retention caused by a mid-intronic substitution (c. 3603 + 2053 G>C, 21st bp of CE), which constructed an exon splicing enhancer [20]. Finally, Zaum et al. reported a patient with two single nucleotide variations flanking exactly the inserted region of the CE in 2017 [11]. Here we report that a new mid-intronic variation (c. 4518 + 512 T>A, 4th bp of CE) generates two ESEs which activate the potential splicing sites then retain the intronic fragment by enhancing the splicing strength.
According to the “frame-shift” hypothesis, reading frame disruption leading to a complete loss of functional dystrophin in muscle causes DMD, and in-frame variants that allow production of an altered but partially functional dystrophin protein causes BMD. When a mutation is out-of-frame, a BMD can still be observed if a residual amount of normal transcript is present (patient II-1 family 1). The CEs retained from pre-mRNA in our cases are separately 58, 167, and 172 bps which are not divisible by 3, so they finally disrupt the reading frame. By skipping the CE, we may correct the cryptic splicing, and generate enough normal transcripts. The US Food and Drug Administration has approved five ASOs for treating genetic diseases, including Vitravene for cytomegalovirus retinitis, Kynamro for familial hypercholesterolemia, Exondys51 for DMD, Spinraza for spinal muscular atrophy, and Tegsedi for hereditary transthyretin amyloidosis [21,22,23,24]. By influencing the SREs, Spinraza causes the SMN2 mRNA to retain the 7th exon, and Exondys51 induces skipping of the 51th exon in the dystrophin transcript. As with Exondys51, other therapeutic molecules such as SRP-4045 and SRP-4053 reframe the dystrophin mRNA transcripts by exon skipping and produce truncated dystrophin with partial function. In contrast, we skip the retained CE to restore the normal transcripts, and generate full-length dystrophin.
Our study emphasizes the need to combine WES and RNA-based methods to detect the variant in the very large DMD gene in which the mutational spectrum is complex. We confirmed the conclusion that a single nucleotide substitution can result in intronic sequence retention by generating new ESEs, and demonstrated that the mis-splicing can be rectified by morpholino modified ASOs. Compared with exon-skipping therapies, CE-skipping therapies represent more personalized, targeted treatment with private variants. However, considering ethical and financial issues, it is difficult to develop clinical trials for private variant at this stage. Next, we will construct an animal model harboring this variant to establish treatment with ASOs.
References
Hoffman EP, Brown RH, Kunkel LM. Dystrophin—the protein product of the Duchenne muscular-dystrophy locus. Cell. 1987;51:919–28.
Birnkrant DJ, Bushby K, Bann CM, Apkon SD, Blackwell A, Brumbaugh D, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and neuromuscular, rehabilitation, endocrine, and gastrointestinal and nutritional management. Lancet Neurol. 2018;17:251–67.
Sansović I, Barišić I, Dumić K. Improved detection of deletions and duplications in the DMD gene using the multiplex ligation-dependent probe amplification (MLPA) method. Biochemical Genet. 2013;51:189–201.
Ankala A, da Silva C, Gualandi F, Ferlini A, Bean LJH, Collins C, et al. A comprehensive genomic approach for neuromuscular diseases gives a high diagnostic yield. Ann Neurol. 2015;77:206–14.
Hua YM, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. Plos Biol. 2007;5:729–44.
Mazzone E, Martinelli D, Berardinelli A, Messina S, D’Amico A, Vasco G, et al. North Star Ambulatory Assessment, 6-minute walk test and timed items in ambulant boys with Duchenne muscular dystrophy. Neuromuscul Disord. 2010;20:712–6.
Cartegni L, Wang JH, Zhu ZW, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–71.
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67.
Hua YM, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82:834–48.
Bonilla E, Samitt CE, Miranda AF, Hays AP, Salviati G, Dimauro S, et al. Duchenne muscular-dystrophy - deficiency of dystrophin at the muscle-cell surface. Cell. 1988;54:447–52.
Zaum AK, Stuve B, Gehrig A, Kolbel H, Schara U, Kress W, et al. Deep intronic variants introduce DMD pseudoexon in patient with muscular dystrophy. Neuromuscul Disord. 2017;27:631–4.
Okubo M, Minami N, Goto K, Goto Y, Noguchi S, Mitsuhashi S, et al. Genetic diagnosis of Duchenne/Becker muscular dystrophy using next-generation sequencing: validation analysis of DMD mutations. J Hum Genet. 2016;61:483–9.
Ikezawa M, Minami N, Takahashi M, Goto Y, Miike T, Nonaka I. Dystrophin gene analysis on 130 patients with Duchenne muscular dystrophy with a special reference to muscle mRNA analysis. Brain Dev. 1998;20:165–8.
Ikezawa M, Nishino I, Goto Y, Miike T, Nonaka I. Newly recognized exons induced by a splicing abnormality from an intronic mutation of the dystrophin gene resulting in Duchenne muscular dystrophy. Mutations in brief no. 213. Online. Hum Mutat. 1999;13:170.
Tuffery-Giraud S, Saquet C, Chambert S, Claustres M. Pseudoexon activation in the DMD gene as a novel mechanism for Becker muscular dystrophy. Hum Mutat. 2003;21:608–14.
Beroud C, Carrie A, Beldjord C, Deburgrave N, Llense S, Carelle N, et al. Dystrophinopathy caused by mid-intronic substitutions activating cryptic exons in the DMD gene. Neuromuscul Disord. 2004;14:10–8.
Gurvich OL, Tuohy TM, Howard MT, Finkel RS, Medne L, Anderson CB, et al. DMD pseudoexon mutations: Splicing efficiency, phenotype, and potential therapy. Ann Neurol. 2008;63:81–9.
Meregalli M, Maciotta S, Angeloni V, Torrente Y. Duchenne muscular dystrophy caused by a frame-shift mutation in the acceptor splice site of intron 26. Bmc Med. Genet. 2016;17:55.
Khelifi MM, Ishmukhametova A, Van Kien PK, Thorel D, Mechin B, Perelman S, et al. Pure intronic rearrangements leading to aberrant pseudoexon inclusion in dystrophinopathy: a new class of mutations? Hum Mutat. 2011;32:467–75.
Trabelsi M, Beugnet C, Deburgrave N, Commere V, Orhant L, Leturcq F, et al. When a mid-intronic variation of DMD gene creates an ESE site. Neuromuscul Disord. 2014;24:1111–7.
Sharma VK, Sharma RK, Singh SK. Antisense oligonucleotides: modifications and clinical trials. Medchemcomm. 2014;5:1454–71.
Syed YY. Eteplirsen: First Global Approval. Drugs. 2016;76:1699–704.
Corey DR. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat Neurosci. 2017;20:497–9.
Keam SJ. Inotersen: First Global Approval. Drugs. 2018;78:1371–6.
Acknowledgements
The authors wish to thank the patients and their families for their participation. JM wants to thank the patient and support from Ni Xie for these years.
Funding
This study was supported by the National Nature Science Foundation of China (81870902) and the Joint Funds for the innovation of science and Technology, Fujian province (2018Y91030173).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Jin, M., Li, JJ., Xu, GR. et al. Cryptic exon activation causes dystrophinopathy in two Chinese families. Eur J Hum Genet 28, 947–955 (2020). https://doi.org/10.1038/s41431-020-0578-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-0578-z
This article is cited by
-
Functional analysis of variants in DMD exon/intron 10 predicted to affect splicing
Journal of Human Genetics (2022)