Abstract
Osteoporosis and obesity are two severe complex diseases threatening public health worldwide. Both diseases are under strong genetic determinants as well as genetically correlated. Aiming to identify pleiotropic genes underlying obesity and osteoporosis, we performed a bivariate genome-wide association (GWA) meta-analysis of hip bone mineral density (BMD) and total body fat mass (TBFM) in 12,981 participants from seven samples, and followed by in silico replication in the UK biobank (UKB) cohort sample (N = 217,822). Combining the results from discovery meta-analysis and replication sample, we identified one novel locus, 17q21.31 (lead SNP rs12150327, NC_000017.11:g.44956910G > A, discovery bivariate P = 4.83 × 10−9, replication P = 5.75 × 10−5) at the genome-wide significance level (ɑ = 5.0 × 10−8), which may have pleiotropic effects to both hip BMD and TBFM. Functional annotations highlighted several candidate genes, including KIF18B, C1QL1, and PRPF19 that may exert pleiotropic effects to the development of both body mass and bone mass. Our findings can improve our understanding of the etiology of osteoporosis and obesity, as well as shed light on potential new therapies.
Similar content being viewed by others
Introduction
Obesity, which has become a major global public health problem [1], is associated with an increased risk of type 2 diabetes, hypertension, cardiovascular disease and so on. In addition, obesity is recognized as the second highest risk factor for cancer [2], which has become one of the potential risks of disability and death worldwide. Obesity is injuring ~65% of adults in the US population [3] and incurring a direct cost of $100 billion per year [4]. Body mass index (BMI, kg/m2), defined as body weight divided by height squared, is the mostly used measure of obesity. Though BMI is widely studied, it is not an ideal phenotype to measure obesity. However, it is less representative of obesity than body fat mass (BFM), which directly associates to the adverse effect of obesity. BFM is a highly heritable trait with heritability estimate of 60% [5].
Osteoporosis, which is characterized by low bone mineral density (BMD) and bone strength, is a common metabolic bone disorder. Osteoporosis can lead to increased risk of fractures, and result in increased morbidity and mortality [6]. It is estimated that over 200 million people are suffering from osteoporosis worldwide. Therefore, osteoporosis has become an alarming health problem [7]. BMD is the standard predictor of osteoporosis and osteoporotic fracture risk. According to the previous study, BMD is also a highly heritable trait with heritability ranging from 0.5 to 0.8 [8].
Previous studies demonstrated that osteoporosis was significantly associated with obesity. Osteoblast cells and adipocyte cells share the same progenitor, bone marrow mesenchymal stem cells, and can trans-differentiate into each other [9]. Adipocytes secrete factors important to bone remodeling, such as estrogen synthesis enzyme, aromatase, and proinflammatory cytokines [10]. Correlation between fat mass and BMD was observed [11]. Considering the complex connection and correlation between osteoporosis and obesity, pleiotropic genes may exist to influence the risks of both diseases.
Genome-wide association studies (GWASs) and their meta-analysis are powerful approaches to identify common variants associated with complex traits [12,13,14]. A number of studies have identified several genomic regions shared by obesity and osteoporosis [15], providing further support for the existence of pleiotropic genes for the two diseases. Importantly, identification of such pleiotropic genes in humans may offer novel insights into the pathogenic links between obesity and osteoporosis. However, loci with pleiotropic effects to both traits are large unknown.
In the present study, aiming to identify pleiotropic loci jointly regulating the development of obesity and osteoporosis, we performed a bivariate GWAS meta-analysis of hip BMD and total body fat mass (TBFM) in seven cohorts and followed by in silico replication in the UK biobank (UKB) cohort sample.
Materials and methods
Discovery sample
We conducted a GWAS meta-analysis of hip BMD and TBFM in seven samples from different ancestries. Three samples were from the in-house studies and the other four were accessed through the database of genotype and phenotype (dbGAP) [16]. All samples were approved by the respective institutional ethics review boards, and all participants provided informed consent before being enrolled into the study. Details of the samples were described previously [17, 18]. Briefly, the first sample comprised 1000 unrelated subjects of European ancestry from the Omaha osteoporosis study (OOS). The second sample comprised 2286 unrelated subjects of European ancestry from the Kansas City osteoporosis study (KCOS). The third sample comprised 1627 unrelated subjects of Chinese Han ancestry from the China osteoporosis study (COS). The other four samples were accessed through the dbGAP. The fourth sample was from the Framingham heart study (FHS). The FHS study is a longitudinal and prospective cohort comprising more than 16,000 related participants spanning three generations of European ancestry [19]. We identified a total of 5800 genotyped and phenotyped FHS participants for analysis. Both the fifth and sixth samples were from the Women’s health initiative (WHI) observational study. The WHI is a partial factorial randomized and longitudinal cohort with over 12,000 genotyped women aged 50–79 years, of African–American or Hispanic ancestry [20]. The sixth sample comprised 850 subjects of African–American ancestry (WHI-AA), and the sixth sample comprised 446 subjects of Hispanic ancestry (WHI-HIS). The last sample, Indiana fragility study (IFS), is a cross-sectional cohort comprising 1493 premenopausal sister pairs of European ancestry [21]. All samples are population-based and their detailed description, including ancestry, gender composition, et al., are shown in Table 1.
Phenotype measurements and modeling
Hip BMD and TBFM were measured by body scan with DXA bone densitometer (Lunar Corp., Madison, WI, USA or Hologic Inc., Bedford, MA, USA) in all the seven samples, following the manufacturer’s protocols. In all samples, covariates, including gender, age, age squared, height (in case of TBFM), height squared (in case of TBFM), and the first five principal components derived from genome-wide genotype data [22], were screened for significance with the step-wise linear regression model implemented in the stepAIC function in R package MASS. Raw TBFM and BMD were adjusted by significant covariates, and the residuals were then normalized by inverse quantiles of standard normal distribution. Normalized residuals were used for subsequent association analysis.
Genotyping and quality control
All GWAS samples were genotyped by high-throughput SNP genotyping arrays (Affymetrix Inc., Santa Clara, CA, USA; or Illumina Inc., San Diego, CA, USA within individual samples), following the manufacturer’s protocols. Quality control (QC) within each sample was implemented at both individual and SNP levels. At the individual level, sex compatibility was checked by imputing sex from X-chromosome genotype data with PLINK [23]. Individuals of ambiguous imputed sex or of imputed sex inconsistent with reported sex were removed. At the SNP level, SNPs violating the Hardy–Weinberg equilibrium rule (P < 1.0 × 10−5) and those containing a minor allele frequency (MAF) <0.01 were removed. Population outliers were monitored by genotype-derived principal components, and outliers were removed if present.
Genotype imputation
GWAS samples were imputed into the 1000 genomes project phase 3 sequence variants (as of May 2013) (see URLs) [24]. Haplotypes representing 240 individuals of European ancestry, 244 of East Asian ancestry, 319 of African ancestry, and 170 of admixed American ancestry were downloaded from the project download site. Haplotypes of bi-allelic variants, including SNPs and bi-allelic insertions/deletions (INDELs), were extracted to form reference panels for imputation. As a QC procedure, variants with zero or one copy of minor alleles were removed.
Each GWAS sample was imputed by the respective reference panel with the closest ancestry. Specifically, the OOS, KCOS, FHS, IFS, and WHI-HIS sample were imputed by the reference panel with European ancestry; the COS sample was imputed by the reference panel with East Asian ancestry and the WHI-AA sample were imputed by the reference panel with African ancestry.
Prior to imputation, a consistency test of allele frequency between the GWAS and reference samples was examined with the chi-square test. To correct for potential mis-strandedness, GWAS SNPs that failed the consistency test (P < 1.0 × 10−6) were transformed into the inverse strand. SNPs that again failed the consistency test were removed from the GWAS sample. Imputation was implemented with FISH [25] to predict missing genotypes.
Association test in individual samples
Each GWAS sample was tested for association between normalized phenotype residuals and genotyped and imputed genotypes under an additive mode of inheritance. Both univariate and bivariate association tests were performed in each individual sample. For unrelated samples, association was examined by the univariate/bivariate linear regression model. For the familial samples (FHS and IFS), a univariate/bivariate mixed linear model was used to account for genetic relatedness within each pedigree [26]. All the analyses were performed by the in-house program GAP, as described previously [27].
Ancestry-specific, trans-ancestry, and sex-stratified meta-analyses
Summary statistics of associations from each GWAS were combined to perform univariate or bivariate meta-analysis. Firstly, we conducted the European ancestry-specific meta-analysis by jointing four European samples (OOS, KCOS, FHS, and IFS). Then, we combined all the seven samples and conducted trans-ancestry meta-analysis and sex-stratified meta-analysis. As a QC procedure, only common or less common (MAF > 0.01 in the European population) and well-imputed (r2 > 0.3 in at least two samples) SNPs were included for analysis.
Both univariate and bivariate meta-analyses were performed under the fixed-effects model [28]. Briefly, for a particular SNP, let (β1i, β2i) be the regression coefficients for the two traits in the ith study (i = 1, …, n, n = 7), and let \({\mathbf{V}}_i = \left[ {\begin{array}{*{20}{c}} {v_{11}} & {v_{12}} \\ {v_{21}} & {v_{22}} \end{array}} \right]\) be the corresponding symmetric variance-covariance matrix for the two regression coefficients. Both β1i, β2i and Vi are obtained from individual study analysis. Define the following data structure:
where B is the vector of regression coefficient, X is the design matrix, and V is the variance-covariance matrix for all studies, respectively.
The generalized least-squared estimator \({\hat{\mathbf{\beta }}}\) of overall regression coefficients is given by
\({\hat{\boldsymbol{ \beta }}} = {\mathbf{(X}}^{\prime} {\mathbf{V}}^{{\mathrm{ - 1}}}{\mathbf{X)}}^{{\mathrm{ - 1}}}{\mathbf{X}}^{\prime} {\mathbf{V}}^{{\mathrm{ - 1}}}{\mathbf{B}}\), which has a normal distribution with mean β and covariance matrix Σ given by
Under the null hypothesis of no association to either phenotype, that is, β = 0 (for both traits), the score statistic
will asymptotically follow a chi-squared distribution with 2 degrees of freedom.
The two univariate test-statisitics are constructed similarly. Specifically,
where \({\hat{\boldsymbol{\beta }}}_1\) and \({\hat{\boldsymbol{\beta }}}_2\) are two elements in \({\hat{\boldsymbol{\beta }}}\), and Σ11 and Σ22 are two diagonal elements in Σ. Under the null hypothesis of no univariate association, that is, β1 = 0 or β2 = 0, T1 or T2 will follow a chi-squared distribution with 1 degree of freedom.
The above meta-analysis model was implemented in an in-house java program BiMeta.jar, which is provided in Supplementary File 1. To monitor potential genetic heterogeneity, the I2 from the two univariate association results was reported.
To test for significant difference of identified loci between men and women, we used a two-sample t-test:
where βM and βF are the β coefficients in men and women, respectively, and SEM and SEF were the standard errors of the β coefficients in men and women, respectively [29].
Sensitivity analysis was performed by removing each study in turn and performing the same meta-analysis in the remaining samples.
Definition of association locus
We conducted univariate and bivariate GWAS meta-analyses of hip BMD and TBFM. Genome-wide significance (GWS) threshold was set to be 5.0 × 10−8. An independent locus was defined as a genomic region of 500 kb to either side of the variant showing the strongest association. A pleiotropic variant is defined based on its GWS in bivariate analysis and nominal significance (P < 0.05) in both univariate analyses.
Cis-expression quantitative trait loci (cis-eQTL) analysis
To investigate the association between the identified SNP and their nearby gene expressions, we performed cis-eQTL analysis. Cis-eQTL information is available for over 50 tissues from the Qtlizer (see URLs), we explored three tissues linked to our traits, including subcutaneous adipose, osteoblasts, and whole blood. We analyzed all lead SNPs and their proxies with strong LD pattern (r2 > 0.8) for their cis-eQTL activity. Briefly, the cis-eQTL was defined as an SNP within 100 kb upstream and downstream of a gene [30]. Using a Bonferroni correction, we set the significance threshold, α, to be 0.05/Ntissue/NSNP.
In silico replication
We further replicated the significant SNPs identified in the discovery sample in the UKB sample [31]. In brief, the UKB sample is a large prospective cohort study of ~500,000 participants from across the United Kingdom, aged between 40 and 69 at recruitment. Ethics approval for the UKB study was obtained from the North West Centre for Research Ethics Committee (11/NW/0382), and informed consent was obtained from all participants. This study used the data requested under the UKB application number 41542, which was covered by the general ethical approval for the UKB study.
Genome-wide genotypes for all subjects were available at 784,256 genotyped autosome markers, and were imputed into UK10K haplotype, 1000 Genomes project phase 3 and Haplotype Reference Consortium reference panels. Subjects who were genotyped but not imputed or who withdraw their consents were removed. All the included subjects are those who were genetically determined as white.
Heel BMD (data field 3148) as evaluated by quantitative ultrasound speed of sound and broadband ultrasound attenuation was used for replicating hip BMD because of the large number of phenotyped subjects and moderately correlated with hip BMD (r = 0.52) [32]. TBFM (data field 23100) as evaluated by the bioelectrical impedance analysis was used. Phenotype modeling for both BMD and BFM was similar to that in the discovery samples, with the exception that both phenotypes were mandatorily adjusted by the top 10 principal components, to adjust for potential population structures. Association was again examined by the univariate/bivariate linear regression model.
Functional annotation
We annotated the functional relevance of the identified SNPs (bivariate P < 5 × 10−8) with HaploReg v4.1 (see URLs) [33]. HaploReg annotates SNPs into different functional categories according to the information from a variety of large experiment projects. These categories include conservation sites, DNase hypersensitivity region, transcription factor binding sites, promoter, enhancer, and others. We annotated lead SNPs and their proxies with strong LD pattern (r2 > 0.8).
For candidate genes, we annotated them by constructing gene interaction networks with STRING (see URLs) [34]. STRING uses information based on gene co-expression, text-mining, and others, to construct gene–gene interaction networks.
Results
European ancestry-specific GWAS meta-analysis
A total of 10,070 participants from four samples were included in the European ancestry-specific GWAS meta-analysis. After QC, a total of 2,374,420 SNPs were included in the meta-analysis. We identified 50 variants at the GWS level, mapping to 18 independent loci (Supplementary Table 1).
Trans-ancestry GWAS meta-analysis
A total of 12,981 participants from seven samples were included in this meta-analysis. Basic characteristics of the discovery samples are listed in Table 1. Sixty-seven percent of the participants are women. The significant covariates for each sample are shown in Supplementary Table 2.
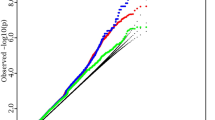
The Manhattan plot of the GWAS meta-analyses is displayed in Fig. 1. In the univariate GWAS meta-analysis of hip BMD, 99 variants were identified at the GWS level, mapping to eight distinct loci: 2q34 (lead SNP rs1429891, NC_000002.12:g.208510208C > A, P = 5.03 × 10−9), 4p16.3 (rs3755955, NC_000004.12:g.1000625G > A, P = 3.89 × 10−9), 6q25.1 (rs10656721, NC_000006.12:g.151691712_151691713insTT, P = 1.97 × 10−9), 7q31.31 (rs58981560, NC_000007.14:g.121382563_121382567del, P = 1.96 × 10−8), 7q21.3 (rs1917486, NC_000007.14:g.96612291C > T, P = 4.72 × 10−8), 8q21.3 (rs536316182, NC_000008.11:g.87846458T > C, P = 1.87 × 10−11), 8q24.12 (rs4876868, NC_000008.11:g.118914750A > G, P = 1.21 × 10−10) and 16q24.1 (rs71390846, NC_000016.10:g.86681109G > C, P = 2.66 × 10−8). Six of the eight loci (4p16.3, 6q25.1, 7q31.31, 7q21.3, 8q24.12, and 16q24.1) are reported to be associated with BMD in previous studies (LD r2 > 0.5 at same region). The remaining two loci, 2q34 and 8q21.3, are novel associated loci for hip BMD.
GWAS −log10 P values are plotted for all SNPs across chromosomes 1–22. Dotted line denotes genome-wide significance level (GWS, 5.0 × 10−8). The red dots denote the variants reaching at GWS level in bivariate GWAS analysis, the blue dots denote the variants reaching at GWS level in univariate GWAS analysis of hip BMD and the green dots denote the variants of TBFM.
In the univariate analysis of TBFM, six variants from the three distinct loci are significant at the GWS level: 1p32.2 (lead SNP rs10789019, NC_000001.11:g.56332765G > A, P = 3.80 × 10−8), 1q31.1 (rs12090181, NC_000001.11:g.190710852T > A, P = 4.49 × 10−8), and 16q12.2 (rs56137030, NC_000016.10:g.53791993G > A, P = 4.72 × 10−8). Although these loci have not been reported to be associated with BFM, two of them, 1q31.1 and 16q12.2, were reported to be associated with BMI, implying that they are known obesity risk variants. The remaining locus at 1p32.2 locus has not been reported to be associated with either BMI or BFM traits. The results of univariate GWAS is shown in Supplementary Tables 3 and 4.
In the bivariate GWAS analysis, three of the above 11 (8 + 3) loci (2q34, 6q25.1, and 8q21.3) remain significant at the GWS level. Among them, the lead SNPs at 2q34 and 8q21.3 are identical in both bivariate and univariate analyses. Interestingly, the lead SNP rs35883965 at 6q25.1 (P = 9.39 × 10−9) is independent of the lead SNP rs10656721 identified in the univariate analysis of hip BMD (LD r2 = 0.006).
In addition to the above three loci, the bivariate analysis also identified 11 loci at the GWS level that are not identified in either univariate analysis: 3q22.1 (rs1389271, NC_000003.12:g.131066262G > T, bivariate P = 1.03 × 10−9), 4q28.2 (rs12512163, NC_000004.12:g.129257175G > A, P = 5.84 × 10−9), 8q21.3 (rs536316182, P = 5.62 × 10−12), 10p15.1 (rs2892347, NC_000010.11:g.6796887G > T, P = 3.43 × 10−10), 10q23.1 (rs7907900, NC_000010.11:g.85607638C > T, P = 1.90 × 10−8), 13q21.31 (rs11840815, NC_000013.11:g.63534155C > A, P = 6.63 × 10−10), 14q32.33 (rs72714919, NC_000014.9:g.104040120G > A, P = 3.12 × 10−9), 15q26.2 (rs10520797, NC_000015.10:g.95647253G > A, P = 1.43 × 10−8), 17q12 (rs2158239, NC_000017.11:g.37448456T > G, P = 1.66 × 10−10), 17q21.31 (rs12150327, NC_000017.11:g.44956910G > A, P = 4.83 × 10−9), 17q21.32 (rs7406170, NC_000017.11:g.48583715G > C, P = 2.19 × 10−8), and 18p12.21 (rs78202598, NC_000018.10:g.13852512G > A, P = 9.92 × 10−9).
In summary, we identified 14 loci in the bivariate meta-analysis. All of the 14 lead SNPs are nominally significant (P < 0.05) in both univariate analyses, implying their potential pleiotropic effect. The results of bivariate GWAS is listed in Supplementary Table 5.
Among the 14 loci, 6q25.1 has been reported to be associated with BMD [35], and 17q21.32 was reported to be associated with obesity-related traits [36]. The remaining 12 loci have not been reported for either BMD or obesity traits.
Sex-stratified meta-analyses
A higher hip BMD and lower TBFM were observed in males for all the four discovery samples (Table 1). We tested whether the 14 pleiotropy lead SNPs identified in the discovery samples showed sex-specific difference (Supplementary Table 6). One SNPs (rs12150327) showed significant gender differences on hip BMD, and three SNPs (rs1389281, rs12512163, and rs78202598) showed significant gender difference on TBFM, indicating these loci are risk only in women/men (Supplementary Table 6).
Follow-up analyses for the 14 loci
For all of the 14 identified lead variants, we performed cis-eQTL analysis in subcutaneous adipose tissue, osteoblast tissue, and whole blood of the GTEx project datasets. Potential candidate genes identified in this analysis included GRN (Granulin Precursor), CCDC170 (Coiled-Coil Domain Containing 170), PLCD3 (Phospholipase C Delta 3), and HOXB2 (Homeobox B2). SNP rs12150327 was the eQTL for both GRN and PLCD3 genes, but the association for GRN is much stronger. The results of cis-eQTL analysis is listed in Supplementary Table 7.
The 14 identified loci were divided into two groups according to their effect direction on both traits: one group containing eight lead SNPs whose effect directions were consistent to both traits, and the other group containing six lead SNPs whose effect directions were opposite. The connection of nearby genes to both traits at these loci was evaluated. Gene was assigned to a locus if the lead SNP is located within the primary transcript, 35 kb upstream or 10 kb downstream of the gene. Protein–protein interaction networks and functional enrichment analysis were performed on the assigned genes using STRING and PathCards (see URLs). A total of 15 and 5 genes were divided into the consistent group and the opposite group, respectively (Supplementary Table 8). Some genes may have functional relevance to both bone and adiposity development. For example, HOXB4 gene at 17q21.32 participates in enbryonic skeletal system morphogenesis. In a microarray-based gene expression analysis, its expression was lower in osteoarthritis patients than in normal controls [37]. In addition, HOXB4 is also an adipocyte regulator which was upregulated at day 11 of adipocyte development [38].
We have also explored the association of the identified loci with six anthropometric traits (including BMI, height, weight, waist circumstance, hip circumstance, and waist hip rate (WHR)) and five bone-related disease traits (including bone disorder, osteoarthritis, osteomyelitis, osteoporosis, and osteopenia) with Gene ATLAS (see URLs), as listed in Supplementary Table 9. At the stringent significance level accounting for multiple testing (α = 0.05/14/11 = 3.25 × 10−4), three SNPs (rs35883965, rs11840815, and rs12150327) were associated with at least one trait. Interestingly, we found that rs12150327 was significantly associated with up to four obesity traits (weight, waist circumstance, hip circumstance, and WHR), strengthening the confidence toward its role in body fat development.
In silico replication in the UKB sample
The 14 loci identified in the discovery sample were subjected to replication in the UKB sample. The lead SNPs rs35883965 at 6q25.1 and rs151106622 (NC_000008.11:g.87914965A > G) at 8q21.3 were not found in the imputed UKB genotypes and were replaced by the second lead SNP rs4869744 (NC_000006.12:g.151586877T > C, bivariate P = 9.54 × 10−9, LD r2 = 0.99) and rs151106622 (bivariate P = 1.42 × 10−8, LD r2 = 0.20), respectively. One other lead SNP rs11840815 at 13q12.31 was not available nor there was suitable proxy SNP in this region, therefore could not be replicated.
The detailed in silico replication results are listed in Supplementary Table 10. In the univariate analysis, five SNPs (rs35883965, rs2158239, rs12150327, rs7406170, and rs78202598) are nominally significant (P < 0.05) for BMD, and one SNP rs12150327 is significant for TBFM (P = 1.04 × 10−4). In the bivariate analysis, two SNPs rs35883965 and rs12150327 are nominally significant. While rs12150327 is nominally significant in both univariate analyses, rs35883965 is significant only for BMD.
Taken together, one SNP rs12150327 is successfully replicated for its pleiotropic effect to both BMD and BFM traits. The main results for rs12150327 is listed in Table 2.
Sensitivity analysis
rs12150327 is significant in the KCOS sample (P = 1.18 × 10−11), but is not significant in any of the other samples. Therefore, we performed a sensitivity analysis by excluding KCOS sample and recalculating the pooled effect size. Results shows the association become nonsignificant (P = 0.963) after excluding the KCOS, indicating that the association is only existence in KCOS sample.
Functional annotation
We annotated the replicated lead variant rs12150327 and its neighboring SNPs (LD r2 > 0.8) through HaploReg. rs12150327 is an expression quantitative trait locus (eQTL) site associated with the target genes CCDC130, FAM187A (Family With Sequence Similarity 187 Member A), DBF4B (DBF4 Zinc Finger B), DCAKD (Dephospho-CoA Kinase Domain Containing), and EFTUD2 (Elongation Factor Tu GTP Binding Domain Containing 2) [39, 40]. It is predicted to have enhancer activity by chromatin states, H3K4me1 and H3K27ac marks. It also has promoter activity suggested by H3K4me3 and H3K9ac markers. All of its ten neighboring SNPs are eQTL sites associated with different genes and all have promoter activity and enhancer activity by different histone marks. Among them, rs75726889 (NC_000017.11:g.44970863G > A, 1.4 kb apart, r2 = 1.0) has promoter activity and enhancer activity in adipose cells and osteoblast cells.
Newly identified locus 17q21.31 and relevant genes
rs12150327 at 17q21.31 is a common (MAF = 0.07 in European population) and imputed SNP with high imputation certainty (r2 > 0.9 in discovery samples). Allele G at this SNP is associated with increased hip BMD (meta-analysis β = 0.07) and decreased TBFM (β = −0.06). A regional plot of rs12150327 is displayed in Fig. 2. It is located between the 5′- of KIF18B (Kinesin Family Member 18B, 9.2 kb) gene and 3′- of C1QL1 (Complement C1q Like 1, 2.8 kb) gene region. Hence, we explored the functional relevance of KIF18B, C1QL1, and the other five eQTL target genes (CCDC103, FAM187A, DBF4B, DCAKD, and EFTUD2) by STRING. The protein–protein interaction network identified dozens of genes that are connected at least one of the seven target genes. Among them are those playing regulatory role in either bone or fat development, such as KIF18B, DBF4B, and EFTUD2 (Fig. 3). Besides, the RACGAP1 (Rac GTPase-activating protein 1) gene is a predicted functional partner of KIF18B, which may contribute to obesity by participating in the regulation of growth-related processes in adipocytes and myoblasts [41]. The PRPF19 (Pre-MRNA Processing Factor 19) gene is a common predicted functional partner of DBF4B and EFTUD2. It participates in the biogenesis of lipid droplets, which may play a role in lipid metabolism and storage [42].
The figure was plotted by STRING. a RACGAP1 gene is a predicted functional partner of KIF18B, which may have possible mechanism contributing to obesity; b PRPF19 gene is a common predicted functional partner of DBF4B and EFTUD2, which may have possible mechanism contributing to obesity. The key genes were marked in black boxes.
Discussion
In the present study, we have performed a bivariate GWAS meta-analysis of hip BMD and TBFM in seven samples to identify the pleiotropy genes which joint regulate the development of obesity and osteoporosis. The significant association signals identified in the discovery sample were further replicated in the UKB sample. Combining the results from discovery and replication samples, one novel locus 17q21.31 was identified to exert pleiotropic effects to both BMD and TBFM.
Many studies have demonstrated the association between 17q21.31 and different traits, such as Alzheimer’s disease [43], breast carcinoma [44], multiple sclerosis [45], and so on, suggesting a complex etiology in this region. Other variants in this region were also reported to be associated with BMD (rs12326005, NC_000017.11:g.45010860G > A, rs4436817, NC_000017.11:g.45014355C > A, rs8071429, NC_000017.11:g.45051538T > A, and rs111443054, NC_000017.11:g.45411797_45411798AC [7]) [14, 36, 46]. However, those lead SNPs are in weak LD (r2 < 0.1) with the one identified in this study.
rs12150327 is located between the KIF18B gene and C1QL1 gene. There is no known role for KIF18B in either bone or fat development. However, one of its predicted functional partners, RACGAP1, is involved in the regulation of growth-related processes in adipocytes and myoblasts. C1QL1 is a Complement 1q (C1q)-related factor. It may regulate the number of excitatory synapses that are formed on hippocampus neurons. C1q plays a key role in activating the classical pathway by recognizing immune complexes [47]. Osteoclasts can produce complement C1Q, which in turn significantly increase the expression of osteoclast genes [48]. Moreover, the absence of C1q/TNF-related protein 9 (CTRP9) can increase food intake in mice, due in part to the upregulated expression of hypothalamic orexigenic neuropeptides. Mice lacking CTRP9 were obese and gained significant weight when fed standard laboratory foods [49]. rs12150327 is also an eQTL variant associated with five different target genes [39, 40]. With STRING, we found that the PRPF19 gene, which is a common predicted functional partner of DBF4B and EFTUD2 gene, may play a role in the biogenesis of lipid droplets. These functional evidences, together with the significant association signals, may suggest the potential etiology of KIF18B gene, C1QL1 gene, and PRPF19 gene in the bone and fat-related biology.
The relationship between obesity and osteoporosis is far more complex than a simple positive or negative correlation [9, 10]. On the one hand, higher body weight exerts a pressure on the bones which increases BMD, which is why obesity was for a long time considered a protective factor against osteoporosis. On the other hand, bone and fat cells share their progenitor, and studies have shown that obesity may negatively affect bone health by the infiltration of adipocytes in the bones. Therefore, we suspect that due to different mechanisms, these pleiotropic loci have opposing effects on BMD and BFM. In this study, we divided the 14 loci into two groups according to their effect direction on both traits, and explored the possible mechanisms of these loci and BMD and BFM, respectively. In the consistent group, we found some functional genes such as HOXB4 that together increase or decrease BMD and BFM, which may support the standpoint that higher body weight exerts a pressure on the bones to increase BMD. However, we did not find any possible mechanisms for the opposite contribution to BMD and BFM in opposite group, although the contribution of these genes to a trait were found. TADA2A gene is a prospective regulator of de novo hepatic lipogenesis in chicken [50]. In addition, TADA2A gene participates in the HATs acetylate histones pathway, and KAT2A and KAT2B genes participate in this pathway too with close relationship to TADA2A. Both KAT2A and KAT2B are required for growth and differentiation of craniofacial cartilage and bone in both zebrafish and mice [51].
Traditional GWAS analysis focuses on the exploration of possible genetic risk loci regulating a single trait, which may lose some valuable information, such as pleiotropic genetic loci. Therefore, we carried out a bivariate GWAS analysis, and expanded the sample size through meta-analysis, so that the statistics power is greatly improved. In addition, BMI is not an idea phenotype for obesity because weight is made up of various body components, including BFM, lean mass, bone mass, and other soft tissues. BFM is the only component that reflects obesity. We used TBFM as a phenotypic trait for obesity rather than BMI, which can strongly explain the association between probable locus and obesity. Moreover, we adopted UKB sample, the current largest research sample, as replication sample, which can significantly improve the reliability of our results. The genetic correlation between heel BMD and hip BMD is reported to be 0.52 [32]. BMD at different skeletal sites share common genetic determinants [52]. Therefore, heel BMD could partially replicate the findings identified in other skeletal sites including hip.
However, there are still some limitations in this study. Firstly, in the discovery stage, the identified SNP rs12150327 is significant only in KCOS sample. The variant become not significant after removing KCOS sample, although the heterogeneity effect was not significant in both univariate meta-analyses (I2 = 29.55 and 11.22 for hip BMD and TBFM, respectively), which indicating that the variant has pleiotropy effect only in some population rather than the entire European ancestry or all human races. Secondly, due to meta-analysis combines samples from different ethnics, it is difficult to explain the population specificity of the associated loci. The purpose of including cross-ethnic samples is to maximize the statistical power of association test, under the assumption that the phenotypic traits of different ethnic groups may have a common genetic basis.
In conclusion, by performing a bivariate GWAS meta-analysis and in silico replication in the large-scale UKB sample, we identified one novel pleiotropy locus at 17q21.31 for hip BMD and TBFM. Our findings can provide insights into genetic pleiotropy underlying osteoporosis and obesity, and can improve our understanding of the etiology of bone and fat development, and shed light on potential new therapies.
Data availability
The GWAS summary statistics were deposited in the GWAS catalog: (ftp://ftp-private.ebi.ac.uk/SummaryStatsUploads/XintongWei_prePMID/); The 1KG phase 3 reference panel can be download at: (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/). Online tools: Qtlizer: (http://genehopper.de/qtlizer); HaploReg v4.1: (http://pubs.broadinstitute.org/mammals/haploreg/haploreg.php); STRING: (https://string-db.org/); Gene Atlas: (http://geneatlas.roslin.ed.ac.uk/); PathCards: (https://pathcards.genecards.org/).
References
Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–81.
Venniyoor A. The most important questions in cancer research and clinical oncology-Question 2-5. Obesity-related cancers: more questions than answers. Chin J Cancer. 2017;36:18.
Flegal KM, Carroll MD, Ogden CL, Johnson CL. Prevalence and trends in obesity among US adults, 1999-2000. JAMA. 2002;288:1723–7.
Wolf AM, Colditz GA. Current estimates of the economic cost of obesity in the United States. Obes Res. 1998;6:97–106.
Elder SJ, Roberts SB, McCrory MA, Das SK, Fuss PJ, Pittas AG, et al. Effect of body composition methodology on heritability estimation of body fatness. Open Nutr J. 2012;6:48–58.
Ramirez-Salazar EG, Carrillo-Patino S, Hidalgo-Bravo A, Rivera-Paredez B, Quiterio M, Ramirez-Palacios P, et al. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene. 2018;679:19–27.
Lampropoulos CE, Papaioannou I, D’Cruz DP. Osteoporosis-a risk factor for cardiovascular disease? Nat Rev Rheumatol. 2012;8:587–98.
Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:303–26.
Kokabu S, Lowery JW, Jimi E. Cell fate and differentiation of bone marrow mesenchymal stem cells. Stem Cells Int. 2016;2016:3753581.
Magni P, Dozio E, Galliera E, Ruscica M, Corsi MM. Molecular aspects of adipokine-bone interactions. Curr Mol Med. 2010;10:522–32.
Ho-Pham LT, Nguyen UD, Nguyen TV. Association between lean mass, fat mass, and bone mineral density: a meta-analysis. J Clin Endocrinol Metab. 2014;99:30–8.
Lu Y, Day FR, Gustafsson S, Buchkovich ML, Na J, Bataille V, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun. 2016;7:10495.
Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–9.
Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2019;51:258–66.
Liu YZ, Pei YF, Liu JF, Yang F, Guo Y, Zhang L, et al. Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PLoS ONE. 2009;4:e6827.
Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, et al. NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–9.
Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei YF, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23:1923–33.
Pei YF, Hu WZ, Yan MW, Li CW, Liu L, Yang XL, et al. Joint study of two genome-wide association meta-analyses identified 20p12.1 and 20q13.33 for bone mineral density. Bone. 2018;110:378–85.
Mahmood SS, Levy D, Vasan RS, Wang TJ. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383:999–1008.
Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials. 1998;19:61–109.
Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
Zhang L, Pei YF, Fu X, Lin Y, Wang YP, Deng HW. FISH: fast and accurate diploid genotype imputation via segmental hidden Markov model. Bioinformatics. 2014;30:1876–83.
Zhang L, Li J, Pei YF, Liu Y, Deng HW. Tests of association for quantitative traits in nuclear families using principal components to correct for population stratification. Ann Hum Genet. 2009;73:601–13.
Zhang L, Pei YF, Li J, Papasian CJ, Deng HW. Univariate/multivariate genome-wide association scans using data from families and unrelated samples. PLoS ONE. 2009;4:e6502.
Konstantopoulos S. Fixed and mixed effects models in meta-analysis. IZA Discussion Paper No. 2198. 2006.
Tin A, Marten J, Halperin Kuhns VL, Li Y, Wuttke M, Kirsten H, et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat Genet. 2019;51:1459–74.
Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779.
Graafmans WC, Van Lingen A, Ooms ME, Bezemer PD, Lips P. Ultrasound measurements in the calcaneus: precision and its relation with bone mineral density of the heel, hip, and lumbar spine. Bone. 1996;19:97–100.
Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–81.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52.
Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–65.
Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104:65–75.
Mucientes A, Herranz E, Moro E, Lajas C, Candelas G, Fernandez-Gutierrez B, et al. Differential expression of HOX genes in mesenchymal stem cells from osteoarthritic patients is independent of their promoter methylation. Cells. 2018;7:244.
Watts KL, Delaney C, Humphries RK, Bernstein ID, Kiem HP. Combination of HOXB4 and Delta-1 ligand improves expansion of cord blood cells. Blood. 2010;116:5859–66.
Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11.
Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43.
Kawashima T, Hirose K, Satoh T, Kaneko A, Ikeda Y, Kaziro Y, et al. MgcRacGAP is involved in the control of growth and differentiation of hematopoietic cells. Blood. 2000;96:2116–24.
Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, et al. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J Biol Chem. 2007;282:2456–65.
Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–17.
Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017;551:92–4.
International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium, Sawcer S, Hellenthal G, Pirinen M, Spencer CC, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9.
Kim SK. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS ONE. 2018;13:e0200785.
Schumaker VN, Zavodszky P, Poon PH. Activation of the first component of complement. Annu Rev Immunol. 1987;5:21–42.
Teo BH, Bobryshev YV, Teh BK, Wong SH, Lu J. Complement C1q production by osteoclasts and its regulation of osteoclast development. Biochem J. 2012;447:229–37.
Wei Z, Lei X, Petersen PS, Aja S, Wong GW. Targeted deletion of C1q/TNF-related protein 9 increases food intake, decreases insulin sensitivity, and promotes hepatic steatosis in mice. Am J Physiol Endocrinol Metab. 2014;306:E779–90.
Desert C, Baeza E, Aite M, Boutin M, Le Cam A, Montfort J, et al. Multi-tissue transcriptomic study reveals the main role of liver in the chicken adaptive response to a switch in dietary energy source through the transcriptional regulation of lipogenesis. BMC Genomics. 2018;19:187.
Sen R, Pezoa SA, Carpio Shull L, Hernandez-Lagunas L, Niswander LA, Artinger KB. Kat2a and Kat2b acetyltransferase activity regulates craniofacial cartilage and bone differentiation in zebrafish and mice. J Dev Biol. 2018;6:27.
Pei YF, Liu L, Liu TL, Yang XL, Zhang H, Wei XT, et al. Joint association analysis identified 18 new loci for bone mineral density. J Bone Min Res. 2019;34:1086–94.
Acknowledgements
We appreciate all the volunteers who participated into this study. YFP and LZ were partially supported by the funding from national natural science foundation of China (31771417 and 31571291). RH was supported by the Medical Health Research Program of Inner Mongolia Autonomous Region Health Commission (201702180). HWD and HS were partially supported by the National Institutes of Health (R01 AR069055, U19 AG055373, R01 MH104680, R01 AR059781, and P20 GM109036), the Franklin D. Dickson/Missouri Endowment and the Edward G. Schlieder Endowment. This study was benefited from a project funded by the Priority Academic Program Development (PAPD) of Jiangsu higher education institutions. The numerical calculations in this paper have been done on the supercomputing system of the National Supercomputing Center in Changsha. The funders had no role in study design, data collection and analysis, results interpretation, or preparation of the manuscript. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Whole Body and Regional Dual X-ray Absorptiometry (DXA) dataset was provided by NIH grants R01 AR/AG 41398. The datasets used for the analyses described in this manuscript were obtained from dbGaP through dbGaP accession phs000342.v14.p10. The WHI program is funded by the National Heart, Lung, and Blood Institute, National 20 Institutes of Health, U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by the Women’s Health Initiative (WHI), and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. Funding for WHI SHARe genotyping was provided by NHLBI Contract N02-HL-64278. The datasets used for the analyses described in this manuscript were obtained from dbGaP through dbGaP accession phs000200.v10.p3. Funding support for the Genetic Determinants of Bone Fragility (the Indiana fragility study) was provided through the NIA Division of Geriatrics and Clinical Gerontology. Genetic Determinants of Bone Fragility is a genome-wide association studies funded as part of the NIA Division of Geriatrics and Clinical Gerontology. Support for the collection of datasets and samples were provided by the parent grant, Genetic Determinants of Bone Fragility (P01-AG018397). Funding support for the genotyping which was performed at the Johns Hopkins University Center for Inherited Diseases Research was provided by the NIH NIA. The datasets used for the analyses described in this manuscript were obtained from dbGaP through dbGaP accession phs000138.v2.p1.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wei, XT., Feng, GJ., Zhang, H. et al. Pleiotropic genomic variants at 17q21.31 associated with bone mineral density and body fat mass: a bivariate genome-wide association analysis. Eur J Hum Genet 29, 553–563 (2021). https://doi.org/10.1038/s41431-020-00727-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-00727-3