Abstract
In this study, we performed a spinal muscular atrophy carrier screening investigation with NGS-based method. First, the validation for NGS-based method was implemented in 2255 samples using real-time PCR. The concordance between the NGS-based method and real-time PCR for the detection of SMA carrier and patient were up to 100%. Then, we applied this NGS-based method in 10,585 self-reported normal couples (34 Chinese ethnic groups from 5 provinces in South China) for SMA carrier screening. The overall carrier frequency was 1 in 73.8 (1.4%). It varied substantially between ethnic groups, highest in Dai ethnicity (4.3%), and no significant difference was found between five provinces. One couple was detected as carriers with an elevated risk of having an SMA affected baby. The distribution of SMN1:SMN2 genotype was also revealed in this study. Among the individuals with normal phenotype, the exon 7 copy-number ratio of SMN1 to SMN2 proved the gene conversion between them. With NGS-based method, we investigated SMA carrier status in Chinese population for the first time, and our results demonstrated that it is a promising alternative for SMA carrier screening and could provide data support and reference for future clinical application.
Similar content being viewed by others
Introduction
Spinal muscular atrophy (SMA) is a rare neuromuscular disease, which is characterized by loss of α motor neurons and progressive degeneration. It is the second most frequent lethal autosomal recessive disease worldwide, often causing death in early life. In the general population, SMA has an incidence of 1/10,000 to 1/5000, and 1 in 35 to 1 in 50 of overall carrier frequency [1,2,3]. According to the onset age and severity of muscle weakness, SMA can be divided into four subtypes. SMA was reported to be caused by the survival motor neuron 1 gene (SMN1), which is located in 5q13 (telomeric form) [4, 5]. More than 95% of the SMA patients are caused by exon 7 deletion of SMN1 gene [6]. A homologous gene, SMN2 (5q13, centromeric form) [4, 7] can play a modifier role on SMA with the its copy number change, positively correlated with a milder symptom [8,9,10].
As is known, carrier screening is a powerful genetic method to prevent the transmission of autosomal recessive or X-linked genetic disorders between generations. The screening of Tay–Sachs disease in the Ashkenazi Jewish population first proved the power of carrier screening. The incidence reduced by more than 90% after several decades’ implementation [11]. Carrier screening of SMA for preconceptional couples and all reproductive age women has been recommended by both American College of Obstetricians and Gynecologists [12] and American College of Medical Genetics and Genomics [6]. Its positive impact on reducing affected births has already been demonstrated [3, 13]. Carrier screening of SMA has been reported in China. For example, Chan et al. performed SMA screening on 569 normal individuals from Hong Kong by real-time quantitative PCR [14]. In Taiwan, 2262 out of 107,611 pregnant women were identified to be with one copy of the SMN1 gene using denaturing high-performance liquid chromatography (DHPLC) or multiplex polymerase chain reaction [3]. Ninety out of 4719 pregnant women from Shanghai were detected to be with only one copy of the SMN1 gene using DHPLC technique [15]. In addition, SMA carrier screening has ever been implemented on hundreds of healthy adults from Guangzhou with multiplex ligation-dependent probe amplification (MLPA) method [16]. However, the populations of these researches targeted only specific regions or specific ethnic groups in China.
In the early stages, SMA carrier screening is performed by traditional methods, such as MLPA, quantitative real-time polymerase chain reaction (QPCR), and DHPLC due to technically challenging [3, 13, 17,18,19]. These technologies are relatively laborous, expensive, and may be inferred by genetic variations located in probe binding sites. Even in the NGS-based expended carrier screening (ECS), traditional tests have to be supplemented additionally to analyze SMA [20,21,22], which are laborious and increase the costs of ECS. Fortunately, NGS-based method has been developed recently for detecting SMN1 and SMN2 copies number [23], making NGS-based ECS of SMA testing with no additional cost.
As is known, China is a vast and multiethnic country. The population mobility, cultural diversity, and geographical distribution of the population makes it difficult to carry on pan-ethnic carrier screening of SMA in China. The study of SMA carrier screening based on pan-ethnic large population is essential by high-throughput test methods, such as NGS, to assess SMA rates for large population and multiethnic groups in Chinese population.
In this study, we first validated the accuracy of the NGS-based method for SMA test, then we investigated SMA carrier rate distribution among 10,585 healthy couples from 34 Chinese ethnic groups in southern China. The results obtained demonstrated that NGS is a promising alternative for SMA carrier screening, and offered a theoretical basis for general carrier screening of SMA in Chinese population.
Materials and methods
Study population
First, in order to demonstrate the accuracy of the NGS-based method in SMA screening, SMN1 copy-number analysis was implemented by both NGS-based method and real-time TaqMan PCR with 2255 samples simultaneously. All the 2255 samples used for validation were collected from May 2014 to December 2017, some of which were with self-identified family history of SMA. For SMA carrier screening, a total of 21,170 samples (10,585 couples) without self-identified family history were analyzed in this study. These couples were initially collected for the Hemoglobinopathies Research Project [24]. Informed consent was required for all the participants at the time of sample collection. All the participants were informed and agreed that the data can be used for other research purposes after de-identification. SMA screening investigation in this study was performed based on the raw sequencing data of the Hemoglobinopathies Research Project [24]. The results of SMA testing were not informed to the participants due to the implementation of this study 3 years after sample collection. This study and all the protocols were approved by the ethics committee of BGI (NO. BGI-IRB 18034).
Targeted region capture and NGS
A previously designed capture array (NimbleGen, Roche) was used to capture the SMN1 gene and other 12 genes selected for carrier testing [25]. The enrichment of target regions was performed according to the previously described protocol [25]. Briefly, genomic DNA extracted was fragmented with sonication, and then was ligated to adapter oligonucleotides (Illumina). DNA libraries were then generated after amplification. The hybridization and elution of DNA libraries with capture probes were performed according to Nimblegen’s standard procedures (Roche NimbleGen, Inc.), followed by sequencing on the HiSeq 2000 or 4000 (Illumina, San Diego, USA).
Data analysis
The primary data were acquired after image processing, error analyses, and base calling according to the Illumina Pipeline (version 1.8) in a run. Then, unqualified reads and adapter sequences were removed to obtain clean reads. The alignment of clean reads against the human genome (version hg19/NCBI37 version) was performed with Burrows-Wheeler Aligner software package (http://bio-bwa.sourceforge.net/). Picard MarkDuplicates (http://broadinstitute.github.io/picard/) was used to mark duplicate reads. Then GATK was then used to do local realignment and re-correction of base quality. The depth of exons and SNP for each sample was generated with GATK Depth Of Coverage. Here, the exon is numbered as the traditional exon numbering (exons 1, 2a, 2b, and 3–8) for gene SMN1 and SMN2 [26, 27], while the reference sequence used for SMN1 and SMN2 is NM_000344.3 and NM_017411.3, respectively (exons 2 and 3 are referred to as 2a and 2b). SMN1 and SMN2 copy-number analysis were conducted using a previously described protocol [28] with a slight modification. The copy number was calculated according to the reads number covering three distinguished base pairs (NC_000005.9:g.70247773C>T, NC_000005.9:g.70247724, and NC_000005.9:g.70247921).
Samples were categorized based on the corresponding credible interval [28]: “likely” carriers (1 copy of SMN1), “unlikely” carriers (≥2 copies of SMN1), and “possibly” carriers (1.5 copies of SMN1, gray area) that be recommended to verify with MLPA in the clinic. All “possible” carriers were recommended to be verified by MLPA.
SMN1 copy-number analysis by real-time TaqMan PCR
After NGS-based SMA testing, the results of 2255 samples were validated using methods other than NGS to demonstrate the accuracy and sensitivity of NGS-based method. SMN1 copy number was detected with real-time TaqMan PCR following the previous description [29]. The primers designed according to two genes’ differentiated nucleotide in exon 7 (c.840C>T, NC_000005.9:g.70247773C>T) for SMN1 exon 7 is 5′-TTCCTTTATTTTCCTTACAGGGTGTC-3′ and 5′-GCTGGCAGACTTACTCCTTAATTTAA-3′, and 5′-TTCCTTTATTTTCCTTACAGGGTGTT-3′ and 5′-GCTGGCAGACTTACTCCTTAATTTAA-3′ for SMN2 exon 7. The deletion range quantified for SMN1 is NC_000005.9:g.70247774_70247809 del, and NC_000005.9:g. 69372354_69372389 del for SMN2.
A commercially available MLPA kit P060-B2 (MRC Holland) was used to quantify SMN1 copy number according to manufacturer’s directions (www.mlpa.com). The probe sequences for SMN1 exon 7 were 5′-TTTTTTTAACTTCCTTTATTTTCCTTACAGGGTTTC-3′ and 5′-AGACAAAATCAAAAAGAAGGAAGGTGCTCACATTCCTTAAATTAAGGA-3′, 5′-GTGAGCACCTTCCTTCTTTTTGATTTTGTCTA-3′ and 5′-AAACCCTGTAAGGAAAATAAAGGAAGTTAAAAAAAATAGCTA-3′ for SMN2 exon 7. The deletion range tested for SMN1 is NC_000005.9:g.70247738_70247821del, and NC_000005.9:g. 69372353_69372352 del for SMN2.
Results
Validation study
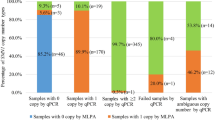
SMN1 copy-number analysis was implemented by the NGS-based method and real-time TaqMan PCR with 2255 samples simultaneously (Fig. 1a). One sample was detected with 0 copy of SMN1 with 2 methods, 69 samples were identified with 1 copy of SMN1, and 10 samples were in the “gray area” by NGS-based method. The remaining were found to be with ≥2 copies of SMN1.
For the ten samples detected as “possibly” carriers by NGS-based method who felled into the “gray area,” three samples (3/10) were verified as carriers (1 copy of SMN1) with real-time PCR, while seven samples (7/10) were found to be with two copies of SMN1. However, no sufficient samples were left to perform MLPA validation for these ten samples. As for the 2175 samples detected to be with ≥2 copies of SMN1 by NGS-based method, 2173 (2173/2175) samples were verified by real-time PCR, only 2 (2/2,175) samples were identified as carriers (1 copy of SMN1) with real-time PCR. And this inconsistency generated by the two methods was further validated with MLPA. As a result, the two samples were also detected to be with two copies of SMN1 by MLPA. As a result, the concordance between the NGS-based method and real-time PCR for the detection of SMA carrier and patient were up to 100% (Fig. 1a).
Carrier burden
The 10,585 couples (21,170 individuals) were from different areas of 5 provinces in Southern China (Supplementary Table S1), and their average age was 29 years old. In total, 10,131 couples (95.71%) provided ethnic information, 326 couples had a racial identity just for 1 partner, 128 couples were without ethnic information. Totally, 34 Chinese ethnic groups were involved. The Han ethnicity was the largest: 14,251 individuals were from the Han ethnic group, comprising 67.32% of all the samples. The remaining were the minority groups and the unknown for ethnic group, accounting for 29.92% and 2.76%, respectively (Supplementary Table S2).
Among 21,170 samples (10,585 couples), 202 of them failed to meet quality control due to low sequencing depth (the total reads of exon 7 of both SMN1 and SMN2 genes are <200) (Fig. 1b). In the remaining (20,968) samples, 85 samples were identified as “possibly” carriers (1.5 copies SMN1, gray area), which were not verified with methods other than NGS due to insufficiency of the samples. The remaining 20,883 samples were included in the followed analysis. Among the 20,883 samples, 10,309 were couples and 265 were single. Of the 20,883 individuals tested, 283 (1.4%) were identified with 1 copy of SMN1 (Fig. 1b). In our cohort, one couple was detected as carriers with an elevated risk of having an SMA affected baby. Theirs SMN2 copy number were 2 and 3, respectively. In the following carrier frequencies analysis categorized by ethnicity and regions, the 85 “possibly” carriers were eliminated.
Carrier frequencies categorized by ethnicity
A total of 20,883 samples (34 self-reported Chinese ethnic groups) were included in the following analysis. The Han ethnicity consist of the largest population (n = 14,117, 67.6%). The other minorities and individuals without a reported ethnic group account for 30.1% (n = 6286) and 2.3% (n = 480), respectively (Table 1). The Zhuang, Hui, Miao, Yi, Tujia, Dong, Buyei, Yao, Bai, and Hani are the top ten minority groups in the south of China [14]. All these groups were included in the present study.
As is shown in Table 1, the overall carrier frequencies between each ethnic group are different. Specifically, individuals of Dai ethnicity showed the lowest carrier frequency (0%), while individuals of Tujia ethnicity showed the highest carrier frequency 4.3%. Both of them were significantly different from other ethnic groups (p < 0.05; Fisher exact test). Besides, two ethnic groups (the Yao and Yi) showed significantly high carrier frequencies (3.5% and 2.5%, respectively; p < 0.05 for both). Of note, the Han ethnicity had an overall carrier frequency of 1.4%. In general, just considering the sample size in this study, there are differences in carrier rates among some ethnic groups. But it is not excluded that these differences may be caused by sample size, thus investigation of large sample size for minority ethnic groups is still necessary.
The carrier frequencies showed slightly different between male and female (Table 1). The overall carrier frequency for female was 1.5%, while for male it was 1.3%. Moreover, the range of carrier frequency was from 0% in Dai to 7.5% in Tujia for female. On the other hand, male carrier frequency was in a narrower range, which was 0% in Dai to 2.1% in Bai. Most of the ethnic carrier frequencies is not statistically specific, no matter considering gender or not.
Carrier frequencies categorized by region
The intra- and interprovincial differences of carrier frequencies were summarized in Table 2. The regional differences were not substantially equivalent among Guangdong, Guangxi, Hainan, Yunnan and Guizhou provinces. Hainan and Yunnan provinces had the highest SMA carrier frequency (1.6%), while the province with the lowest frequency was Guizhou province (1.0%). For different areas of the same province, the difference for carrier rate was significant. For example, it ranged from 0.4% in Tunchang to 2.8% in Qionghai. However, these differences may be caused by insufficient sample size too.
Among the five provinces, the carrier frequencies were different in men and women as well. The lowest female carrier frequency was 1.2% in Guizhou, while the highest one was 1.8% in Yunnan. The range of male carriers was from 0.7% in Guizhou to 1.7% in Hainan. Similar to the situation of the total carrier frequency, interprovincial gender carrier frequencies occupied a larger range (0–2.6% in women; 0–3.1% in men) than intra-provincial ones.
Gene dosage of SMN1 and SMN2
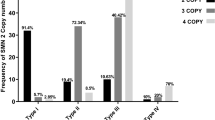
The copy numbers of SMN2 could modulate SMA phenotype. In this study, the relationship between SMN2 and SMN1 gene copy number was analyzed. In the carriers (283/20,883), the SMN2 gene had one to four copies. The majority (91.5%; 19,106/20,883) of healthy individuals carried two copies of wild-type SMN1, and the remaining had three (7%, n = 1434) or four (0.3%, n = 60) copies of the SMN1 gene (Tables 3 and 4). In the two genotypes with more than three copies, at least two SMN1 alleles were on the same chromosome. In the samples with two or three SMN1 alleles, the SMN2 gene copy number varied from 0 to 4. In carriers and normal individuals, the SMN1 copies increase was associated with SMN2 copies reduce. Most SMN1 carriers (83.4%; 236/283) had more than one copy of SMN2, while the portion of samples with more than one copy of SMN2 decreased from 61.3% to 31.7% in a normal population with the increase of SMN1 gene number.
The copy-number ratio of SMN1/SMN2 was available for each ethnic group (Table 3). The majority of ethnic distribution was similar to the overall trend described above, though there were several significantly higher or lower cases. For example, in the normal population with two SMN1 copies, the Han, Miao, and Yi groups showed greater proportion with 2:2, 2:1, and 2:0 copy-number ratio of SMN1/SMN2 (7650/20,883, 292/20,883 and 27/20,883, respectively; p < 0.05 for all). On the other hand, the three groups showed less proportion with 2:0, 2:2, and 2:1 ratio (556/20,883, 323/20,883, and 122/20,883, respectively; p < 0.05 for all). The Yi and Yao groups, showing statistical high carrier frequencies, was significantly high when copy-number ratios of SMN1/SMN2 was 1:3 (6/20,883 and 3/20,883, respectively; p < 0.05 for both). Similarly, the population number of Tujia was high when the ratio was 1:1 (4/20,883; p < 0.05).
The intra- and interprovincial copy-number ratio of SMN1/SMN2 was determined as well (Table 4). Guizhou province had fewer carriers compared with the whole, while the carriers in this province with SMN1/SMN2 ratio of 1:3 (7/20,883; p < 0.05) was significantly lower. In some provinces, only several cities had significantly greater or fewer carriers, like Dongguan and Zhuhai in Guangdong. The proportion with SMN1/SMN2 ratio of 1:2 (2/4755; p < 0.05) was consistent to less carrier in Zhuhai, while the proportion with SMN1/SMN2 ratio of 1:3 (7/4755; p < 0.05) was consistent to the greater carrier in Dongguan.
Discussion
NGS technology is the current trend for clinical carrier testing since it has the ability to determine the status of multiple genetic diseases in parallel. It shows irreplaceable technological advantages over traditional methods, especially in the field of ECS. SMA is a severe and high-incidence genetic disease, it has been recommended for carrier screening. In NGS-based ECS, SMA is included supplemented with traditional technological methods, such as MLPA or real-time TaqMan PCR [20, 21]. The newly developed NGS-based SMA carrier screening makes SMA testing as a part of a comprehensive NGS carrier-testing platform. Previous studies already performed a comparison between sequencing-based and MLPA carrier statuses, which showed a strong correlation [23]. What is more, the NGS validation of phase 3 participants in the 1000 Genomes Project showed that the proportion of SMA carriers closely matched the proportion in subpopulations [28]. In a previous study of the first the NGS-based method for SMA testing, the sensitivity and specificity to detect SMA carriers with one copy of SMN1 was 100% and 99.6%, respectively [23]. The NGS results were confirmed by conventional methods like qPCR or MLPA [23]. In this study, we validated the NGS-based method (with some modification) by real-time PCR, the concordance between the NGS-based method and real-time PCR for the detection of SMA carrier and patient were up to 100%. Our study differentiated the SMN1 and SMN2 copy number in large-scale Chinese population, which makes it possible to perform SMA testing in NGS-based multiple carrier testing. The SMN1 and SMN2 genotypes were also revealed during the test. We were able to differentiate SMA carriers (SMN1/SMN2; 1/1) from noncarriers (2/2 or 3/3), although their SMN1 to SMN2 copy-number ratios were the same.
The results of the screening for SMA carriers from five provinces showed that the carrier frequency was 1 in 73.8 (1.4%) in southern China. It was comparable with SMA carrier frequencies in subpopulation worldwide, which was from 1 in 100 in Black to around 1 in 50 Caucasian and Asian populations [30]. The clear ethnic information in previous studies in China was limited to the Han group or Taiwanese [3, 16, 31]. The pan-ethnic study was still missing, which can represent the diversity of the Chinese population. In our study, certain ethnic groups had significantly higher or lower carrier frequencies (Table 3). One possible explanation of this phenomenon is that the sample size is not large enough in a certain ethnic group. Another explanation may be the intermarry across ethnic groups is still unpopular. For example, all the intermarry ratios of Han and Yi/Yao/Tujia/Dai are below 20%. In this way, the minor ethnic groups keep the population specificity. Although the carrier frequencies might be population-specific in several ethnic groups, the SMA carrier status fell into a certain range across subpopulations. Therefore, it is essential to carry on the pan-ethnic SMA carriers test in China.
Interestingly, the carrier frequency was a little lower than most of the previous studies in China (ranging from 1.6% in Hong Kong to 2.39% in Guangdong province) [3, 14,15,16, 31]. There is only one study in Liuzhou, Guangxi province with a carrier frequency of 1.2%, which was lower than that is in our study [32]. Considering the sample size, our result was more convincing refer to the combined estimated carrier frequency [30]. Compared with previous studies in China, our study was carried on in the larger population (20,883) in broader regions. Therefore, the data with less sample bias were more accurate for estimating the proportion of SMA carriers in China.
The SMN1/SMN2 copy-number information in this cohort provides a closer view of the genotypes in general population. The proportion of samples with three copies of the SMN1 gene or more in minority of healthy individuals (7.3%; n = 1494) was higher than previous observation (about 4%) in normal population [8]. Two SMN1 copies on the same chromosome was detected only when the gene copy number of SMN1 is more than 3. This may be explained by the SMN2 to SMN1 gene conversion. Since the total copy number of SMN1 and SMN2 is consistent in this situation, the SMN2 copy number would decrease after the gene conversion. We showed herein that the correlation of SMN1 copy number increase and SMN2 copy number decrease in the general population, which proved the hypothesis of gene SMN2 to SMN1 conversion. This trend has also been reported in a previous study of gene conversion between SMN1 and SMN2 [33]. The ethnic and geographic distribution of SMN1/SMN2 was not studied because of the limited sample size in previous researches. In this study, our finding showed that the distribution of SMN1/SMN2 gene copy numbers is similar across ethnic groups, though there are several exceptions in both healthy individuals and carriers. When linked with carrier frequencies, copy-number ratios of SMN1/SMN2 can explain part of the ethnic-specific carrier difference. For example, the proportion of 1:3 copy-number ratio of SMN1/SMN2 was significantly higher in the Yi and Yao groups. The distribution across various regions was also indifferent in most cases with only a few exceptions. For example, province Guizhou, with a lower carrier frequency, had fewer carriers with SMN1/SMN2 ratio of 1:3. The reason is not clear, but the carrier screening of SMA is meaningful in a broad region based on this large-scale general population study.
From the view of birth control, there is 1 SMA heterozygous at-risk couple in 10,309 couples, which passed quality control and was in the nongray zone among 10,309 couples analyzed. Since the overall male and female carrier frequencies were 1.3% and 1.5%, respectively, the theoretical risk couple frequency is 1.95‱. The actual frequency (1/10,309; 0.97‱) in our cohort was close to it.
This retrospective study had several drawbacks. For example, the lack of reports for the patients and follow-up data (such as pregnancy outcomes). Due to insufficiency of the samples, the 85 “possibly” carriers (1.5 copies SMN1, gray area) detected by NGS-based method were not validated by MLPA. Some of these “possibly” carriers may also be true SMA carriers. This may possibly result in the lower carrier frequency of our cohort comparing to most of carrier frequencies reported in previous studies [3, 14,15,16, 31]. There’s also a limitation of high-throughput sequencing-based screening, it would miss the following carriers: two functional genes are located on the same chromosome, and a nonfunctional gene is located on the other chromosome. The frequency of this “2 + 0” genotype carrier was 0.11% in the Asian population [34]. Therefore, the observed difference could be reduced. SMA could also be caused by variants in the SMN1 gene. Although we took the point variant in exon 7 and exon 8 into consideration, carriers could have other intragenic variants at the SMN1 gene [1, 35].
In summary, we carried out SMA carrier investigation among 10,585 healthy couples from various ethnic groups in southern China with NGS-based method. The overall SMA carrier rate was 1 in 73.8 (1.4%). The results indicated significant differences between ethnic groups (highest in Dai ethnicity (4.3%), but not between five provinces). In addition, the distribution of SMN1:SMN2 genotype was revealed in this study. Among the individuals with normal phenotype, the exon 7 copy-number ratio of SMN1 to SMN2 proved the gene conversion between them. Our study illustrated that NGS-based method is a promising alternative for SMA carrier screening, and the results provided data support and reference for future clinical application.
Data availability
The information of the variant detected in this study can be found at the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/variation/) with accession number SCV001334438. The identifiable information of the subjects used in this study is kept in our clinical laboratory to protect subjects’ privacy, while their nonidentifiable genomic sequencing data have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb with accession code CNP0000801.
References
Ogino S, Wilson RB. Spinal muscular atrophy: molecular genetics and diagnostics. Expert Rev Mol Diagn. 2004;4:15–29.
Mandelker D, Schmidt RJ, Ankala A, Gibson KM, Bowser M, Sharma H, et al. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet Med. 2016;18:1282–9.
Su YN, Hung CC, Lin SY, Chen FY, Chern JPS, Tsai C, et al. Carrier screening for spinal muscular atrophy (SMA) in 107,611 pregnant women during the period 2005-9: a prospective population-based cohort study. PLoS ONE. 2011;6:e17067.
Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–65.
Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–14.
Prior TW. Carrier screening for spinal muscular atrophy. Genet Med. 2008;10:840–2.
Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AHM, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–8.
McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–22.
Wirth B, Brichta L, Schrank B, Lochmüller H, Blick S, Baasner A, et al. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet. 2006;119:422–8.
Calucho M, Bernal S, Alías L, March F, Venceslá A, Álvarez FJR, et al. Correlation between SMA type and SMN2 copy number revisited: an analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul Disord. 2018;28:208–15.
Kaback MM. Screening and prevention in Tay-Sachs disease: origins, update, and impact. Adv Genet. 2001;44:253–65.
Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–15.
Archibald AD, Smith MJ, Burgess T, Scarff KL, Elliott J, Hunt CE, et al. Reproductive genetic carrier screening for cystic fibrosis, fragile X syndrome, and spinal muscular atrophy in Australia: outcomes of 12,000 tests. Genet Med. 2018;20:513–23.
Chan V, Yip B, Yam I, Au P, Lin CK, Wong V, et al. Carrier incidence for spinal muscular atrophy in southern Chinese. J Neurol. 2004;251:1089–93.
Gong B, Zhang L, Hou YP, Hu HY, Li HC, Tan MY, et al. Carrier screening for spinal muscular atrophy in 4719 pregnant women in Shanghai region. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2013;30:670–2.
Fang P, Li L, Zeng J, Zhou WJ, Wu WQ, Zhong ZY, et al. Molecular characterization and copy number of SMN1, SMN2 and NAIP in Chinese patients with spinal muscular atrophy and unrelated healthy controls. BMC Musculoskelet Disord. 2015;16:11.
Cusco I, Barcelo MJ, Baiget M, Tizzano EF. Implementation of SMA carrier testing in genetic laboratories: comparison of two methods for quantifying the SMN1 gene. Hum Mutat. 2002;20:452–9.
Su YN, Hung CC, Li H, Lee CN, Cheng WF, Tsao PN, et al. Quantitative analysis of SMN1 and SMN2 genes based on DHPLC: a highly efficient and reliable carrier-screening test. Hum Mutat. 2005;25:460–7.
Arkblad EL, Darin N, Berg K, Kimber E, Brandberg G, Lindberg C, et al. Multiplex ligation-dependent probe amplification improves diagnostics in spinal muscular atrophy. Neuromuscul Disord. 2006;16:830–8.
Martin J, Asan, Yi YT, Alberola T, Iglesias BR, Almazán JJ, et al. Comprehensive carrier genetic test using next-generation deoxyribonucleic acid sequencing in infertile couples wishing to conceive through assisted reproductive technology. Fertil Steril. 2015;104:1286–93.
Abulí A, Boada M, Rodríguez‐Santiago B, Coroleu B, Veiga A, Armengol L, et al. NGS-based assay for the identification of individuals carrying recessive genetic mutations in reproductive medicine. Hum Mutat. 2016;37:516–23.
Haque IS, Lazarin GA, Kang HP, Evans EA, Goldberg JD, Wapner RJ. Modeled fetal risk of genetic diseases identified by expanded carrier screening. JAMA. 2016;316:734–42.
Feng Y, Ge X, Meng L, Scull J, Li J, Tian X, et al. The next generation of population-based spinal muscular atrophy carrier screening: comprehensive pan-ethnic SMN1 copy-number and sequence variant analysis by massively parallel sequencing. Genet Med. 2017;19:936–44.
Shang X, Peng Z, Ye Y, Asan, Zhang X, Chen Y, et al. Rapid targeted next-generation sequencing platform for molecular screening and clinical genotyping in subjects with hemoglobinopathies. EBioMedicine. 2017;23:150–9.
Zhao S, Xiang J, Fan C, Asan, Shang X, Zhang X, et al. Pilot study of expanded carrier screening for 11 recessive diseases in China: results from 10,476 ethnically diverse couples. Eur J Hum Genet. 2019;27:254–62.
Bürglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, et al. Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996;32:479–82.
Chen Q, Baird SD, Mahadevan M, Besner-Johnston A, Farahani R, Xuan JY, et al. Sequence of a 131-kb region of 5q13.1 containing the spinal muscular atrophy candidate genes SMN and NAIP. Genomics. 1998;48:121–7.
Larson JL, Silver AJ, Chan D, Borroto C, Spurrier B, Silver LM. Validation of a high resolution NGS method for detecting spinal muscular atrophy carriers among phase 3 participants in the 1000 Genomes Project. BMC Med Genet. 2015;16:100.
Gomez-Curet I, Robinson KG, Funanage VL, Crawford TO, Scavina M, Wang W. Robust quantification of the SMN gene copy number by real-time TaqMan PCR. Neurogenetics. 2007;8:271–8.
Verhaart IEC, Robertson A, Wilson IJ, Aartsma-Rus A, Cameron S, Jones CC, et al. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy—a literature review. Orphanet J Rare Dis. 2017;12:124.
Sheng-Yuan Z, Xiong F, Chen YJ, Yan TZ, Zeng J, Li L, et al. Molecular characterization of SMN copy number derived from carrier screening and from core families with SMA in a Chinese population. Eur J Hum Genet. 2010;18:978–84.
Tan J, Zhang X, Wang Y, Luo S, Yang F, Liu B, et al. Screening for spinal muscular atrophy mutation carriers among 4931 pregnant women from Liuzhou region of Guangxi. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2018;35:467–70.
Ogino S, Gao S, Leonard DG, Paessler M, Wilson RB. Inverse correlation between SMN1 and SMN2 copy numbers: evidence for gene conversion from SMN2 to SMN1. Eur J Hum Genet. 2003;11:723.
Hendrickson BC, Donohoe C, Akmaev VR, Sugarman EA, Labrousse P, Boguslavskiy L, et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J Med Genet. 2009;46:641–4.
Ogino S, Wilson RB, Gold B. New insights on the evolution of the SMN1 and SMN2 region: simulation and meta-analysis for allele and haplotype frequency calculations. Eur J Hum Genet. 2004;12:1015–23.
Acknowledgements
We thank all the blood donors for their invaluable contribution to this study.
Funding
The study was supported by Special Foundation for High-level Talents of Guangdong (2016TX03R171).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhao, S., Wang, W., Wang, Y. et al. NGS-based spinal muscular atrophy carrier screening of 10,585 diverse couples in China: a pan-ethnic study. Eur J Hum Genet 29, 194–204 (2021). https://doi.org/10.1038/s41431-020-00714-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-00714-8
This article is cited by
-
Test development, optimization and validation of a WGS pipeline for genetic disorders
BMC Medical Genomics (2023)
-
Screening and prenatal diagnosis of survival motor neuron gene deletion in pregnant women in Zhaoqing city, Guangdong Province
BMC Medical Genomics (2023)
-
Clinical application value of expanded carrier screening in the population of childbearing age
European Journal of Medical Research (2023)
-
Population WGS-based spinal muscular atrophy carrier screening in a cohort of 1076 healthy Polish individuals
Journal of Applied Genetics (2023)
-
SMA carrier testing using Real-time PCR as a potential preconception screening tool
Egyptian Journal of Medical Human Genetics (2022)