Abstract
Connective tissue disorders are a spectrum of diseases that affect the integrity of tissues including skin, vasculature, and joints. They are often caused by variants that disrupt genes encoding components of extracellular matrix (ECM). The fibulin glycoproteins are ECM proteins important for integrity of tissues including dermis, retina, fascia, and vasculature. The fibulin family consists of seven members (fibulins-1 to -7) and is defined by a fibulin-type domain at the C-terminus. The family is associated with human diseases, for instance a variant in FBLN1, encoding fibulin-1, is associated with synpolydactyly, while one in EFEMP1, encoding fibulin-3, causes Doyne honeycomb degeneration of the retina. Loss-of-function of fibulins-4 and -5 causes cutis laxa, while variants in fibulins-5 and -6 are associated with age-related macular degeneration. Of note, EFEMP1 is not currently associated with any connective tissue disorder. Here we show biallelic loss-of-function variants in EFEMP1 in an individual with multiple and recurrent abdominal and thoracic herniae, myopia, hypermobile joints, scoliosis, and thin translucent skin. Fibroblasts from this individual express significantly lower EFEMP1 transcript than age-matched control cells. A skin biopsy, visualised using light microscopy, showed normal structure and abundance of elastic fibres. The phenotype of this individual is remarkably similar to the Efemp1 knockout mouse model that displays multiple herniae with premature aging and scoliosis. We conclude that loss of EFEMP1 function in this individual is the cause of a connective tissue disorder with a novel combination of phenotypic features, and can perhaps explain similar, previously reported cases in the literature.
Similar content being viewed by others
Introduction
Variants that alter the function of proteins, that comprise or organise components of connective tissue, result in disorders that affect numerous structures including joints, skin, and internal organs. For example, Marfan syndrome, caused by variants in FBN1 (MIM: 134797), results from abnormalities of fibrillin function within the extracellular matrix (ECM), leading to anomalies of the blood vessels, skeleton, and eyes [1, 2]. Ehlers–Danlos syndromes (EDS) affect the skin, joints and vasculature [3]. Some forms of EDS arise as a result of defective collagen [4, 5].
The fibulins are an essential component of connective structures, including the dermis, vasculature, and fascia where they contribute to the elasticity and functional integrity of these tissues [6]. Fibulins are extracellular glycoproteins that consist of a fibulin-type domain at the C-terminus, preceded by repeating calcium-binding epidermal growth factor (EGF)-like modules [7]. The family has seven members with fibulins-1 and -2 defined by the presence of an anaphylatoxin (AT) module at the N-terminus of the protein [6]. Fibulins-3, -4, and -5 belong to a second group of smaller fibulin proteins lacking the AT module. Fibulin-6 is the largest fibulin and has repeating immunoglobulin and thrombospondin domains N-terminal to the EGF domains. Finally, fibulin-7 is structurally similar to fibulins-3, -4, and -5 except for the presence of a unique sushi domain [8].
Fibulin-1 and -2 are expressed in basement membranes and elastic fibres during embryonic development and persist in cardiac and vascular tissues postnatally [9]. The fibulin-1 knockout mouse is embryonic lethal. However, fibulin-2 is dispensable for mouse development suggesting some functional redundancy between the two paralogues [10, 11]. This fibulin subgroup supports extracellular structures by binding fibronectin, tropoelastin, and proteoglycans [7]. Homozygosity for a single variant in FBLN1 (MIM: 135820), encoding fibulin-1, may be causal for a complex type of synpolydactyly [12]. Fibulin-4 and -5 bind to tropoelastin and are critical for the assembly of elastic fibres during development [8]. Variants in EFEMP2 (MIM: 604633) and FBLN5 (MIM: 604580) (encoding fibulins- 4 and -5) result in prominent cutis laxa, while FBLN5 is also associated with age-related macular degeneration (AMD) [13,14,15,16]. Fibulin-6 is expressed in skin fibroblasts and retinal pigment epithelium, while fibulin-7 interacts with fibronectin and heparin and is expressed in the teeth and hair follicles, placenta, and developing cartilage [8]. One missense variant in HMCN1 (MIM: 608548) encoding fibulin-6 (hemicentin), also segregates with AMD in a large family [17].
Fibulin-3, encoded by EFEMP1 (MIM: 601548) is structurally most similar to fibulin-4 (EFEMP2) and fibulin-5 (FBLN5) [18, 19]. EFEMP1 is described as being expressed primarily in the vasculature, retina, and skin fibroblasts [7, 20, 21]. Despite structural similarity to fibulins-4 and -5, fibulin-3 demonstrates little to no interaction with ECM proteins that are known to interact with fibulin-4 and -5 such as fibulin-1, fibronectin, and tropoelastin [7, 19]. Consequently, while variants in EFEMP2 and FBLN5 cause a connective tissue phenotype, no such association has previously been found for EFEMP1 until now [13, 14].
A recurrent pathogenic, missense variant in EFEMP1 (p.(Arg345Trp) (NP_001034437.1)), has previously been implicated in the autosomal dominant disorder Doyne honeycomb (DH) degeneration of the retina, a retinal dystrophy that displays significant phenotypic overlap with AMD [20]. The Efemp1 knockout mouse model, which displays a prominent and striking connective tissue phenotype, demonstrates no sign of a retinal disease, suggesting that the DH degeneration phenotype is unlikely to be conferred by a loss-of-function mechanism [19]. Study of the mouse knockout model demonstrated that complete loss of Efemp1 results in reduced strength and resilience of the fascia, resulting in herniae that became progressively more significant as the animals aged [19].
In this study, we report an individual with a pronounced connective tissue presentation in association with biallelic truncating variants in EFEMP1 (NM_001039348.3: c.[320_324delTGGCA];[615T > A]). We propose that this entity is likely to represent another distinct form of autosomal recessive connective tissue dysfunction with a pronounced visceral, as distinct from cutaneous, presentation.
Materials and methods
Ethics and consent
The individual described in this work consented to participate in a research study under approved protocols MEC/08/08/094 and 13/STH/56 (Health and Disability Ethics Committee, New Zealand) and images of him are also presented with his consent.
Sequence analysis
Exome libraries were constructed and sequenced by Otogenetics (Atlanta, USA) using a HiSeq2500 (Illumina), with prior enrichment using the SureSelect V5 + UTRs (Agilent) capture platform. Sequence reads were paired end, with read lengths of 125 bases. Processing for alignment and standard variant calling was based on GATK Best Practice Guidelines (GATK v3.8) [22, 22]. Paired-end reads in fastq format, derived from a single individual, were aligned to the reference sequence (GRCh37 assembly) using the Burrows–Wheeler Aligner v0.7.17 [24] with the mem algorithm. Duplicate reads were identified using Picard MarkDuplicates (GATK v2.18.11) [25]. Base Quality Score Recalibration was then used to produce the aligned files in bam format. A variant call of single nucleotide variants (SNVs) and short insertions/deletions (indels) was generated using GATK HaplotypeCaller, producing a single-sample GVCF. Joint genotyping of the patient’s GVCF, along with 302 unrelated samples sequenced and analysed identically, was performed using GATK GenotypeGVCFs to produce a standard variant call dataset containing variant information for all samples. This was followed by GATK LeftAlignAndTrimVariants and then Variant Quality Score Recalibration was undertaken independently for SNPs and indels. To facilitate the filtering of patient variants, SnpEff v4.3 [26] was used to annotate with gene context information. Annotation for population allele frequencies was added using BCFtools annotate (v1.9), with data from the gnomAD project (v2.0) [27]. The variants found to be most likely causative of the phenotype in this individual were uploaded to the online database ClinVar.
Cell culture
Dermal fibroblasts from the affected individual and an age-, and sex-matched control were expanded in GIBCO Dulbecco’s modified eagle media (ThermoFisher Scientific) with 15% foetal bovine serum (Moregate, NZ) and 1% Penicillin/Streptomycin (GIBCO, ThermoFisher Scientific) at 37 °C and 5% CO2. Cell lines were treated with cycloheximide (100 µg/ml) (Merck) in DMSO (Merck) or DMSO vehicle control.
RNA extraction and cDNA synthesis
RNA extraction from pelleted fibroblasts was performed using the NucleoSpin© RNA plus RNA isolation kit (Macherey-Nagel, Germany) as per manufacturer’s instructions. RNA was primed for cDNA synthesis with random primers (ThermoFisher Scientific), OligoDT (Sigma custom oligos, Merck) and dNTPs. cDNA synthesis was performed using the Superscript III reverse transcriptase kit (ThermoFisher Scientific).
Quantitative PCR
Primers for gene expression analysis are shown in Table S1 (Sigma custom oligos, Merck). Quantitative PCR (qPCR) was performed using LightCycler 480 SYBR Green master mix 1 (Roche, Germany), on the LightCycler 480 II real-time PCR machine (Roche). Data analysis was performed using the LightCycler 480 software (Roche) and qBase plus 2 (Biogazelle, Belgium).
Vector assembly and sequencing
PCR products were amplified from cycloheximide treated fibroblast cDNA using primers in Table S1 (Sigma custom oligos, Merck) with AmpliTaq® Gold DNA polymerase (ThermoFisher Scientific). PCR products were assembled into the pGEM-T vector using the pGEM-T Easy kit (Promega, WI, USA). Constructs were Sanger sequenced using BigDye terminator reaction mix (Applied Biosystems, CA, USA) and analysed by capillary sequencing on the 3730xl DNA Analyser (ThermoFisher Scientific) by the Otago Genetic Analysis Service (Department of Anatomy, University of Otago).
Histological analysis
Skin biopsies were obtained from scar and healthy tissue on the individual’s shin. Samples were paraffin embedded and 10 nm sections were cut using a microtome. Sections were stained with haematoxylin and eosin (H and E), and Verhoeff–van Gieson (VVG) stains.
Results
Clinical description
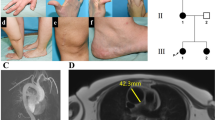
The proband is the only child of non-consanguineous parents of European ancestry. He has two maternal and one paternal half siblings, all in good health. His birthweight was 3.1 kg at 38 weeks gestation. At 6 weeks of age it was noted that he had bilateral inguinal herniae which were repaired only to recur repeatedly over the ensuing two decades. He demonstrated mild motor delays (walked at age 24 months) and hypotonia but no delay in language acquisition. On assessment at age seven he was noted to be dolichocephalic with downslanting palpebral fissures, and to have a high, narrow palate and a long narrow face (Fig. 1a). His carpal bone age was advanced by 1 year. He had a mild pectus excavatum but no scoliosis at that time. He had prominent arachnodactyly and his skin appeared thin and translucent but not wrinkled or loose. Although healing and scar formation were normal, he did tend to bruise easily. His shoulder and knee joints exhibited hypermobility, a property that was less evident in his small joints. His arm span to height ratio was 1.04.
Clinical images of the proband. a Distinctive facial features include a long, narrow face, ectropia of the lower eyelids, and downslanting palpebral fissures; b The skin is thin and translucent; c Large Bochdalek hernia on both sides of the mediastinum with loops of small bowel herniated into the thorax, along with spleen and mesenteric vessels. An obturator hernia is also evident on the left with bladder content; d Diaphragmatic hernia with small bowel loops and stomach in the thorax with multiple pleural based bullae (*) throughout both lung fields; e Coronal MRI of the pelvis with a bladder diverticulum (*) and left obturator hernia (**) containing bladder evident; f Coronal MRI of the pelvis with a pelvic floor hernia with small bowel loop plus bladder content present (***) in addition to a small left obturator hernia
As childhood and early adolescence progressed high myopia was diagnosed although the lenses remained in normal position with no demonstrable abnormalities of the suspensory ligaments or retina. His final height was 203 cm (+3.8 SDS); it is uncertain if his tall stature is partly attributable to familial factors, as a paternal height was not available. His skin has become progressively more lax with age and bilateral ectropia of the lateral portions of the lower eyelids have formed (Fig. 1a, b). He developed the ability to partially sublux his large joints, including his shoulders and knees, at will. His Beighton score was 2/9.
Throughout adolescence and in his early twenties the proband developed multiple complex herniae that were refractory to surgical management. These included complex sciatic and obturator herniae, bilateral Bochdalek herniae (with substantial amounts of abdominal viscera located in the thoracic cavity; Fig. 1c, d), bladder diverticula resulting in recurrent urinary tract infections (Fig. 1e, f), and multiple large pulmonary bullae resulting in haemoptysis and respiratory compromise. An echocardiogram demonstrated no cardiac compromise, dilatation of the aortic root or pulmonary trunk. An MRI of the lumbar spine demonstrated mild dural ectasia. A scoliosis evolved that was concave to the right in the cervicothoracic region and concave to the left in the thoracolumbar region. It did not require surgical treatment.
Measurement of his alpha-1-antitrypsin level demonstrated a normal level and phenotype and plasma homocysteine levels were normal. Sequencing of coding regions of FBN1, CHST14, COL3A1, TGFBR1, TGFBR2, ACTA2, SMAD3, TGFB2, FBN2, EFEMP2, MYH11, NOTCH1, and SLCA10 demonstrated no variants of potential pathogenic significance and hence he was enroled in a study for whole-exome sequencing (WES). A thorough search for disease-causing variants was undertaken of the known causative genes, as listed on OMIM, for EDS (ADAMTS2, ATP7A, CHST14, COL12A1, COL1A1, COL1A2, COL3A1, COL5A1, COL5A2, CRTAP, FKBP14, FLNA, P3H1, PLOD1, SLC39A13) and Cutis Laxa (ALDH18A1, ATP6V0A2, ATP6V1E1, ATP7A, EFEMP2, ELN, FBLN5, LTBP4, PYCR1, RIN2, SLC2A10) using the whole-exome data. No variants of clinical significance were found.
Exome sequencing
The individual’s parents are unaffected, so de novo dominant, autosomal recessive, and X-linked maternal inheritance models were initially considered as potential genetic explanations for his presentation. Due to the lack of parental samples, de novo status for any candidate variant could not be determined limiting the ability to identify causal variants of this nature. To account for this limitation and to consider other inheritance patterns a stringent variant filtering strategy was adopted. A panel of 302 controls was constructed, using individuals with no known relationship to the patient and no similar phenotype (as described in the Methods). Variants in the control panel were excluded from consideration, eliminating platform and pipeline artefacts, as well as removing common variants. After this step 1187 variants remained from a total of 133,994. Variants unique to the patient were then subjected to quality filtering, only retaining those with a read depth ≥8, and where the variant allele represented more than 20% of the total reads at that position. Due to the limitation of interpreting the pathogenic impact of non-coding and missense variants, only high impact variants, absent from gnomAD (141,456 unrelated individuals, with no known severe paediatric disease) were retained, leaving only eight novel variants. The remaining variants were prioritised using probability of loss-of-function (pLI) scores for their cognate genes (gnomAD v2.0). Four variants remained in genes with pLI scores greater than 0.9 - EFEMP1, MAST3, and PKNOX1. All prioritised variants were heterozygous. However, two novel loss-of-function variants were observed in EFEMP1 encoding fibulin-3 (NM_001039348.3: c.[320_324delTGGCA];[615T > A], predicting the protein changes, p.(Met107fs) and p.(Tyr205*) (Fig. 2a). Phasing of the EFEMP1 variants to determine if these variants are in cis or trans was not possible from the exome sequence data due to their distance from one another (>30 kb) and the lack of parental data. The EFEMP1 variants were subject to further experimental analysis based on the remarkably similar phenotype of the Efemp1 knockout mouse [19] and the importance of the fibulin protein family in the integrity of connective tissues [7]. The two variants were submitted to ClinVar and assigned the following accession numbers: NM_001039348.3: c.[320_324delTGGCA]; SCV000995918 and NM_001039348.3: c.[615T > A]; SCV000995919.
a EFEMP1 gene structure (NM_001039348.3), variants discovered in this study are highlighted, numbers represent exons; b qPCR data showed a significant reduction in EFEMP1 expression from affected individual’s fibroblasts compared with those from an age- and sex-matched control. EFEMP1 expression was normalised to GAPDH (***p < 0.001); c Panels 1–4; healthy skin biopsy sections taken from the affected individual, treated with Verhoeff–van Gieson (VVG) to stain the elastin fibres. Elastin is stained black. At ×20 magnification, the elastic fibres appear normal and correctly distributed (arrows). Scale bars represent 100 µm (Color figure online)
Analysis of allele segregation
No paternal sample was available to demonstrate segregation but Sanger sequencing of the available maternal sample demonstrated that the NM_001039348.3: c.[615T > A] variant was inherited from the mother, with the NM_001039348.3: c.[320_324del] absent at her EFEMP1 locus. In order to determine that the EFEMP1 variants were in trans, fibroblasts from the affected individual were treated with 100 ng/ml cycloheximide and RNA extracted and cDNA prepared. A region of EFEMP1 encompassing both variants was PCR-amplified, PCR products were assembled into vectors and individual clones were sequenced (n = 7). Analysis of all clones showed that the two variants are mutually exclusive (Fig. S1) indicating that they are carried on separate alleles. These data indicate that this individual is a compound heterozygote for two high-impact, loss-of-function variants in EFEMP1.
Quantitative measurement of EFEMP1 transcript
To determine if the NM_001039348.3: c.[615T > A];[320_324del] variants decrease gene expression, qRT-PCR (primers: Table S1) was performed from cDNA not treated with cycloheximide extracted from patient skin fibroblasts. The expression of EFEMP1 was greatly diminished to an almost undetectable level, compared with EFEMP1 expression in age-and sex-matched control fibroblasts (~19-fold reduction, p < 0.001) (Fig. 2b). Therefore, the biallelic loss-of-function variants are associated with significantly reduced expression of EFEMP1.
Histological analysis of the skin
Fibulins-4 and -5 are important in the maintenance of dermal structural integrity, as evidenced by the role of these proteins in cutis laxa [13, 14]. An incisional full-thickness skin biopsy was taken from the affected individual for histological assessment and examined under the light microscope. The biopsy revealed a normal epidermis, dermis, and appendages. An elastin stain demonstrated a normal distribution, density, and structure of fibres. Delicate elastin fibres were present in the papillary dermis and thicker fibres present in the reticular dermis (Fig. 2c). Associated blood vessels demonstrated unremarkable elastic laminae.
Discussion
We present here an individual with a previously uncharacterised connective tissue disorder that severely affects the internal viscera and to a lesser extent joint function and spinal alignment. WES revealed biallelic loss-of-function variants in EFEMP1, encoding fibulin-3. Subsequently, we demonstrated that the affected individual has negligible expression of EFEMP1.
A dominantly inherited missense variant in EFEMP1 (p.(Arg345Trp)) has previously been associated with both Malattia Leventinese (ML) and DH retinal dystrophy [20], conditions with a similar phenotype to AMD and no extraocular connective tissue manifestations. There are no recorded cases in the literature of phenotypes associated with recessive, loss-of-function genotypes at EFEMP1 and our individual has no clinical signs of retinal dystrophy. We prioritised the two variants we identified in EFEMP1 because of the occurrence of disease-causing variants in other fibulin family members in the connective tissue disorder, cutis laxa [13, 14] and the phenotype of the Efemp1 knockout mouse (inguinal herniae and pelvic organ prolapse) [19] which shows remarkable similarity to the proband studied here.
Elastic fibres in the skin of the Efemp1−/− mouse appeared slightly reduced when studied under light microscopy with electron microscopy revealing some fragmentation [19]. Similarly, the elastic fibres in the skin of our individual appeared normal under light microscopy. However, the elastic fibres in the internal fascia of the mouse were virtually absent, suggesting that the core pathogenic element contributing to this aspect of the phenotype is a functional insufficiency of visceral fascia [19]. We do not have access to a sample of fascia from our individual but it is possible that he has a similar, dramatic reduction in elastic fibres, presumably leading to his multiple severe and recurrent herniae. The mouse model also demonstrates other phenotypic features in common with the individual studied here, including scoliosis and reduced body fat [19].
Efemp1 promotes chondrogenesis and is specifically expressed in the superficial zone of articular cartilage in mouse and humans. Efemp1−/− mice have more severe age-related osteoarthritis [28]. No other joint phenotypes are reported in the mouse and therefore the hypermobility phenotype may be unique to humans. It will be important to observe individuals with loss-of-function EFEMP1 for osteoarthritis.
Fibulin-3 is most homologous to fibulins-4 and -5, although fibulin-3 seems to lack the ability to interact with the common binding partners of fibulin-4 and -5, including fibulin-1, fibronectin and tropoelastin [7]. Variants in EFEMP2 and FBLN5 (fibulins-4 and -5) both cause severe recessive forms of cutis laxa along with multiple other connective tissue abnormalities [13, 14]. The affected individual described here has translucent skin without skin overfolding and has remained so, since childhood. Therefore, we have not applied a diagnosis of cutis laxa, although it is acknowledged that there are close phenotypic similarities between this phenotype and other forms of the condition, especially those caused by variants in FBLN5 and ATP6V0A2. It could be the case that fibulins-4 and -5 are crucial for the integrity of the skin, whereas fibulin-3 has a similar role in the fascia and is compensated for by its homologues in the dermis.
Other work has progressed understanding of the retinal dystrophy phenotypes associated with a recurrent variant in EFEMP1 and how it differs from the loss-of-function phenotype we describe here. Fibulin-3 binds strongly to tissue inhibitor of metalloproteinases-3 (TIMP-3), and is thought to support TIMP-3 in remodelling the ECM [29]. Variants in TIMP3 (MIM: 188826) cause another macular degeneration disorder, Sorsby fundus dystrophy (SFD). In SFD, ML, and DH retinal dystrophies, as well as AMD, there is accumulation of TIMP-3 and fibulin-3 between the retinal pigment epithelia and Bruch's membrane [29]. This suggests a gain-of-function mechanism in the retinal dystrophy phenotypes, including the p.(Arg345Trp) variant in EFEMP1. Loss of fibulin-3, therefore, is not associated with retinal dystrophy and instead may lead to myopia.
The individual described here presents with a novel connective tissue disorder that shares numerous features with Marfan syndrome but without the cardiac complications. Two historical reports in the literature describe families with a recessive, connective tissue disorder, resembling Marfan syndrome, that shares considerable phenotypic overlap with our individual [30, 31]. Shared features include inguinal, femoral and Bochdalek herniae, visceral diverticula (colon or bladder), myopia with the absence of lens dislocation, hypotonia, and arachnodactyly. Joint hypermobility and translucent skin seem unique to our individual. Many features shown by the individual described here are also shared with recessive cutis laxa caused by variants in LTBP4 (MIM: 613177 and 604710, respectively). Individuals with homozygous and compound heterozygous variants in LTBP4 have cutis laxa, severe respiratory distress with diaphragmatic hernia, diverticulosis, and joint hypermobility [32]. This condition, therefore, displays great similarity to that which we have described here and perhaps the two share a common biochemical pathway. Other connective tissue disorders share many features of the condition we are presenting here, for example herniation, diverticulae, easy bruising and distinctive facial features are also found in recessive cutis laxa [33] and some forms of EDS [3]. However, the condition presented here appears different because of the extreme herniation phenotype and lack of loose skin. Therefore, in conclusion, we present an individual with a complex connective tissue phenotype with severe disruption to his connective viscera. Two variants were discovered in EFEMP1 that cause near-complete loss of gene expression. This individual’s phenotype may represent a novel connective tissue disorder, more individuals with the same condition and loss-of-function variants in EFEMP1 will be required to confirm this disorder as a unique entity.
Web resources
OMIM: https://omim.org/
ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/
Ensembl: http://asia.ensembl.org/index.html
NCBI: https://www.ncbi.nlm.nih.gov/
GATK best practice guidelines: https://software.broadinstitute.org/gatk/best-practices
BCF tools: https://samtools.github.io/bcftools/bcftools.html
GnomAD: https://gnomad.broadinstitute.org/
SnpEff: http://snpeff.sourceforge.net/
References
Milewicz DM, Pyeritz RE, Crawford ES, Byers PH. Marfan syndrome: defective synthesis, secretion, and extracellular matrix formation of fibrillin by cultured dermal fibroblasts. J Clin Investig. 1992;89:79–86.
Dietz HC, Cutting CR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–9.
Beighton P, Paepe AD, Steinmann B, Tsipouras P, Wenstrup RJ. Ehlers-Danlos syndromes: revised nosology. Am J Med Genet. 1998;77:31–37.
Malfait F, de Paepe A. Molecular genetics in classic Ehlers–Danlos syndrome. Am J Med Genet Part C Semin Med Genet. 2005;139C:17–23.
Pepin M, Schwarze U, Superti-Furga A, Byers PH. Clinical and genetic features of Ehlers–Danlos syndrome type IV, the vascular type. N Engl J Med. 2000;342:673–80.
Chu M-L, Tsuda T. Fibulins in development and heritable disease. Birth Defects Res Part C Embryo Today Rev. 2004;72:25–36.
Kobayashi N, Kostka G, Garbe JHO, Keene DR, Bächinger HP, Hanisch F-G, et al. A comparative analysis of the fibulin protein family: biochemical characterization, binding interactions, and tissue localization. J Biol Chem. 2007;282:11805–16.
de Vega S, Iwamoto T, Yamada Y. Fibulins: multiple roles in matrix structures and tissue functions. Cell Mol Life Sci. 2009;66:1890–902.
Zhang H-Y, Timpl R, Sasaki T, Chu M-L, Ekblom P. Fibulin-1 and fibulin-2 expression during organogenesis in the developing mouse embryo. Dev Dyn. 1996;205:348–64.
Cooley MA, Kern CB, Fresco VM, Wessels A, Thompson RP, McQuinn TC, et al. Fibulin-1 is required for morphogenesis of neural crest-derived structures. Dev Biol. 2008;319:336–45.
Sicot F-X, Tsuda T, Markova D, Klement JF, Arita M, Zhang R-Z, et al. Fibulin-2 Is dispensable for mouse development and elastic fiber formation. Mol Cell Biol. 2008;28:1061–7.
Debeer P, Schoenmakers EFPM, Twal WO, Argraves WS, De Smet L, Fryns JP, et al. The fibulin-1 gene (FBLN1) is disrupted in a t(12;22) associated with a complex type of synpolydactyly. J Med Genet. 2002;39:98–104.
Hucthagowder V, Sausgruber N, Kim KH, Angle B, Marmorstein LY, Urban Z. Fibulin-4: a novel gene for an autosomal recessive cutis laxa syndrome. Am J Hum Genet. 2006;78:1075–80.
Markova D, Zou Y, Ringpfeil F, Sasaki T, Kostka G, Timpl R, et al. Genetic heterogeneity of cutis laxa: a heterozygous tandem duplication within the fibulin-5 (FBLN5) gene. Am J Hum Genet. 2003;72:998–1004.
Lotery AJ, Baas D, Ridley C, Jones RPO, Klaver CCW, Stone E, et al. Reduced secretion of fibulin 5 in age-related macular degeneration and cutis laxa. Hum Mutat. 2006;27:568–74.
Stone EM, Braun TA, Russell SR, Kuehn MH, Lotery AJ, Moore PA, et al. Missense variations in the fibulin 5 gene and age-related macular degeneration. N. Engl J Med. 2004;351:346–53.
Schultz DW, Mahan A, Humpert AJ, Runckel C, Luzier CW, Cassera M, et al. Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet. 2003;12:3315–23.
Zhang Y, Marmorstein LY. Focus on molecules: fibulin-3 (EFEMP1). Exp eye Res. 2010;90:374–5.
McLaughlin PJ, Bakall B, Marmorstein AD, Marmorstein LY, Liu Z, Davis EC, et al. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum Mol Genet. 2007;16:3059–70.
Stone EM, Lotery AJ, Munier FL, Héon E, Piguet B, Guymer RH, et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet. 1999;22:199–202.
Lecka-Czernik B, Lumpkin CK, Goldstein S. An overexpressed gene transcript in senescent and quiescent human fibroblasts encoding a novel protein in the epidermal growth factor-like repeat family stimulates DNA synthesis. Mol Cell Biol. 1995;15:120–8.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491-8.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60.
Van der Auwera G, Carneiro M, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.11–11.10.33.
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92.
Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2015;536:285–91.
Hasegawa A, Yonezawa T, Taniguchi N, Otabe K, Akasaki Y, Matsukawa T, et al. Role of fibulin 3 in aging-related joint changes and osteoarthritis pathogenesis in human and mouse knee cartilage. Arthritis Rheumatol. 2017;69:576–85.
Klenotic PA, Munier FL, Marmorstein LY, Anand-Apte B. Tissue inhibitor of metalloproteinases-3 (TIMP-3) is a binding partner of epithelial growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1): implications for macular degenerations. J Biol Chem. 2004;279:30469–73.
de Silva H, Gunawardena T, Law F. Unusual complications in siblings with marfanoid phenotype. Arch Dis Child. 1996;75:247–8.
Clunie G, Mason J. Visceral diverticula and the Marfan syndrome. Br J Surg. 1962;50:51–52.
Urban Z, Hucthagowder V, Schürmann N, Todorovic V, Zilberberg L, Choi J, et al. Mutations in LTBP4 cause a syndrome of impaired pulmonary, gastrointestinal, genitourinary, musculoskeletal, and dermal development. Am J Hum Genet. 2009;85:593–605.
Hbibi M, Abourazzak S, Idrissi M, Chaouki S, Atmani S, Hida M. Cutis Laxa syndrome: a case report. Pan Afr Med J. 2015;20:3–3.
Acknowledgements
We would like to thank the proband and his family for their agreement to participate in this study, the Histology Department at the University of Otago for technical assistance, Sam Hawarden for his contribution to the whole-exome analysis, and Curekids NZ for funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Driver, S.G.W., Jackson, M.R., Richter, K. et al. Biallelic variants in EFEMP1 in a man with a pronounced connective tissue phenotype. Eur J Hum Genet 28, 445–452 (2020). https://doi.org/10.1038/s41431-019-0546-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-019-0546-7
This article is cited by
-
Fibulin-3 knockout mice demonstrate corneal dysfunction but maintain normal retinal integrity
Journal of Molecular Medicine (2020)