Abstract
The HoxD cluster is critical for vertebrate limb development. Enhancers located in both the telomeric and centromeric gene deserts flanking the cluster regulate the transcription of HoxD genes. In rare patients, duplications, balanced translocations or inversions misregulating HOXD genes are responsible for mesomelic dysplasia of the upper and lower limbs. By aCGH, whole-genome mate-pair sequencing, long-range PCR and fiber fluorescent in situ hybridization, we studied patients from two families displaying mesomelic dysplasia limited to the upper limbs. We identified microduplications including the HOXD cluster and showed that microduplications were in an inverted orientation and inserted between the HOXD cluster and the telomeric enhancers. Our results highlight the existence of an autosomal dominant condition consisting of isolated ulnar dysplasia caused by microduplications inserted between the HOXD cluster and the telomeric enhancers. The duplications likely disconnect the HOXD9 to HOXD11 genes from their regulatory sequences. This presumptive loss-of-function may have contributed to the phenotype. In both cases, however, these rearrangements brought HOXD13 closer to telomeric enhancers, suggesting that the alterations derive from the dominant-negative effect of this digit-specific protein when ectopically expressed during the early development of forearms, through the disruption of topologically associating domain structure at the HOXD locus.
Similar content being viewed by others
Introduction
Hox genes encode transcription factors, which play critical roles during animal embryonic development. In mammals, 39 Hox genes are grouped at four genomic loci, designated as the HoxA to HoxD gene clusters [1]. This distinctive genomic organization is closely associated with a regulatory process referred to as ‘collinearity’, i.e., the tight correspondence that exists between the topological order of these genes within each cluster, on the one hand, and the succession of their expression territories along the anterior–posterior embryonic axis, on the other hand [2,3,4,5]. HoxD genes are essential in the patterning mechanisms controlling vertebrate limb development and they display specific proximal to distal expression patterns that specify the future skeletal elements [6].
In rodents, multiple conserved noncoding sequences (CNS) regulating HoxD genes have been described in both the telomeric and centromeric regions flanking the gene cluster. The centromeric regulatory landscape includes a global control region, several regulatory islands (I–V) dispersed within the Lnp-Atp5g3 gene desert, and the Prox element, which is located between Lnp and Evx2 [4, 7, 8]. These centromeric regulators control the coordinated transcription of Hoxd13 to Hoxd9, Lnp, and Evx2 that will pattern and shape the digits. Recently, two additional enhancers—CNS 39 and 65—located in the opposite (telomeric) gene desert were shown to activate Hoxd9 to Hoxd11 in the incipient limb bud to organize structures in the prospective arm and forearm [9]. Chromosome capture conformation experiments have revealed that these centromeric or telomeric CNS contact various HoxD genes through chromatin loops that activate their transcription [4, 9]. Overall, a model was proposed whereby two subsequent waves of HoxD genes transcription occur, under the control of two different chromatin structures referred to as topologically associating domain (TAD) [10]. Early on, the Hoxd9 to Hoxd11 genes are under the transcriptional control of the telomeric TAD and pattern the proximal part of the limbs leading to the arm and forearm [5]. Subsequently, the Hoxd9 to Hoxd13 genes are switched on under the control of the centromeric TAD thus contributing to digits modeling [9, 11].
Mesomelic dysplasias are a heterogeneous group of inherited skeletal dysplasias characterized by disproportionate shortness of the middle segment of the limbs, most often associated with other skeletal anomalies, and usually involving both upper and lower limbs thus resulting in moderate short stature among them Leri–Weill dyschondrosteosis caused by SHOX haploinsufficiency and its homozygous form, Langer dysplasia, are the most common [12]. In addition to several conditions, recognizable on nonskeletal features, such as Robinow or Ellis–van Creveld syndromes or the more recently delineated Verloes–David–Pfeiffer syndrome, various unclassified forms of unknown cause have been reported, mostly in single families with a unique phenotype including in most cases other skeletal problems [12, 13]. However, a recognizable pattern of skeletal involvement has been described in a few patients, such as in Nievergelt type or Savarirayan type of mesomelic dysplasia.
Among these rare disorders, Kantaputra type mesomelic dysplasia (MDK) has emerged as a distinct autosomal dominant entity, reported to date in four families [14,15,16,17,18]. MDK is characterized by marked mesomelic shortness of the upper and lower limbs associated with carpal and tarsal synostosis of variable severity, distal hypertrophy of fibulas and progressive ankylosis of the proximal interphalangeal joint of the fingers. Microduplications including the HOXD gene cluster were simultaneously identified in two unrelated families with MDK [19, 20].
Mesomelic involvement limited to the upper limbs was described in 1988 by Fryns et al. [21] in a female patient and her father and, subsequently, in 2005 by Mégarbané and Ghanem [22] in a boy whose father showed unilateral involvement. We report here an additional family with the same mesomelic involvement confined to the upper limbs. These three families suggest the existence of a separate autosomal dominant condition consisting of ulnar dysplasia that we propose to term Fryns type mesomelic dysplasia. We performed molecular analyses both in this additional family and in the family reported by Mergarbané and Ghanem and show that this phenotype is caused by an inverted duplication of the HOXD gene cluster. We discuss the potential molecular mechanisms underlying this condition and provide a general framework to account for the effects of genetic rearrangements at this locus based on both the loss- and gain-of-function of various HOXD genes during early limb bud development.
Clinical reports
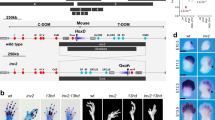
Patient III-2 from family 1 was the second daughter of unrelated parents (Fig. 1a). She was referred for ‘homozygote dyschondrosteosis’. She was born at 37 weeks of gestation by caesarean section. Chromosome analysis on amniotic fluid was performed for intra-uterine growth retardation of unknown cause and was normal 46,XX. Birth length was 41 cm (<3rd percentile), birth weight 1780g (<3rd percentile), and head circumference 32 cm (25th percentile). Physical examination at birth showed short and bowed forearms, ulnar deviation of the hands and mild camptodactylies with no lower limbs anomalies. During follow-up there was a progressive catch up of growth and improvement of camptodactylies. Psychomotor development was normal. On examination at the age of 3 years ½, height was at 97.2 cm (0 SD) with a lower segment at 48 cm, weight at 15 kg (0 SD), and OFC at 51.5 cm (+1.5 SD). She had markedly short and bowed forearms with ulnar deviation of the hands, and mild camptodactylies of the 2nd and 3rd fingers on both side (Fig. 1c). Palmar creases were normal. Shoulders and elbows were normal, whereas wrist flexion–extension and pronation–supination movements were limited. Lower limbs and spine were normal. The patient had no dysmorphic facial features and no palate malformation. Her father (II-1) was similarly affected. His parents and his brother and sister were not affected. Before the birth of patient III-2 he received a diagnosis of dyschondrosteosis and chromosome analysis was normal. Skeletal radiographs of both patients showed severe developmental abnormality of the ulnae, which were extremely short and thick, associated with marked radial bowing but no Madelung deformity (Fig. 1c). The tibiae and fibulae were normal.
Pedigrees of families 1 (a) and 2 (b) described here. Dup/WT: patient with a heterozygous duplication of the HOXD gene cluster; WT/WT: individual with two wild-type HOXD gene clusters; NA not available, mos mosaic, black symbols: phenotypically affected patients; white symbols: unaffected individuals; gray square: affected father with unilateral involvement and a mosaic duplication. c Photographs of the patient III-2 of family 1 at the age of 3 years ½ showing the mesomelic involvement limited to the upper limbs with shortness and bowing of the forearms, ulnar deviation of the hands, and mild camptodactylies. Radiographs of the upper limbs of the patient at the age of 3 years. R right limb, L left limb
Patients II-1 and I-1 from family 2 (Fig. 1b) are a boy and his father who were previously described by Mégarbané and Ghanem [22]. They displayed a similar upper limb involvement although it was unilateral in the father.
Methods
Cytogenetic and aCGH studies
Informed consent for genetic analyses was obtained from the patients according to national ethical guidelines and following the Helsinki Declaration. Rearrangements and phenotypic details were submitted to DECIPHER (https://decipher.sanger.ac.uk/). Karyotyping based on R or G banding was performed using standard methods on metaphase spreads from peripheral blood of the patients. Genomic DNA was extracted from peripheral blood using standard protocols. Array comparative genomic hybridization (aCGH) experiments were performed using Agilent Human Genome CGH 60 K oligonucleotide arrays (Agilent, Santa Clara, CA; www.agilent.com) with the ISCA design (www.iscaconsortium.org) in patients II-1 and III-2 of family 1 or Human Genome CGH 400 K oligonucleotide arrays in patients I-1 and II-1 of family 2 following the protocols provided by Agilent. Subsequently, we used a custom targeted 60 K Agilent array to fine map the breakpoints of the duplications in patient II-1 of family 1. Custom array comprised 7255 probes covering a 1.45 Mb in the 2q31 region including the HOXD gene cluster and had an average resolution of 150 bp. The arrays were analyzed with the Agilent scanner and the Feature Extraction software (v.9.1.3). Graphical overview was obtained using the CGH analytics software (v.3.5.14). Fluorescence in situ hybridization (FISH) was performed in family 1 with a specific probe (RP11-203K19) located in the 2q31 duplicated region and a probe (GS-892G20) located in the 2p subtelomeric region used as control probe. Real-time quantitative PCR (qPCR) with primers designed inside the duplicated regions was used for independent confirmation of the aCGH results and for parental inheritance [23]. Two individuals (NA18542 and NA18968) of Asian origin from the HapMap Project were obtained from the Coriell Institute [IHMC, 2005] and were also analyzed using Agilent Human Genome CGH 60 K oligonucleotide arrays with the custom targeted array.
Breakpoint sequencing
Whole genome mate-paired libraries were prepared with DNAs of patients III-2 of family 1 and II-1 of family 2 according to the Ion Mate-Paired Library Preparation protocol using the 5500 SOLiD mate-paired library kit (Life Technologies, Carlsbad, CA, USA). Briefly, 3 µg of each genomic DNA was sheared to yield 3 kb fragments. Blunt-ended repaired DNA fragments were ligated to MP adapters and circularized. Then, cleavage of circular DNA removed a linear DNA fragment composed of MP adapters and genomic DNA ends. Finally, Ion adapters were ligated to these linear fragments before sequencing. Sequencing was performed on Ion Proton sequencer using Ion PI sequencing 200 kit v3 and Ion PI chip v2 (Life Technologies) with a read length of 200 bases. Sequences were generated with the Torrent suite software v4.0.4 (Life Technologies).
Paired-end whole-genome sequencing was performed from 3 µg genomic DNA of individuals I-1 and I-2 of family 1 and patient II-1 of family 2 according to the Illumina TruSeq DNA PCR-free protocol (Illumina, San Diego, CA, USA) on an Illumina NextSeq 500 following standard protocols. Structural variants and copy-number variants were detected using BreakDancer v 1.4.5 and ERDS v 1.1 and then annotated using Svagga (https://gitlab.inria.fr/NGS/svagga).
Long-range PCR (TaKaRa LA Taq, TAKARA) and direct sequencing (primer sequences are available upon request) were performed to characterize the breakpoints at a molecular level in families 1 and 2.
Fiber FISH
EBV-transformed lymphoblasts of patient II-1 of family 1 were used to perform fiber FISH analyses. Briefly, the cultured cells were pelleted and embedded in 0.5% low melting agarose blocks. Embedded cells were lysed in 1% lithium dodecylsulfate buffer at 37 °C overnight, washed in 0.02% N-Lauroylsarcosine buffer and, next, in Tris EDTA (10:1) solutions at 37 °C during 1 h. Agarose blocks were melted on polylysine glass slides at 55 °C. Chromatin fibers were obtained by stretching the melting blocks with a second slide and cross-linked 5 min under UV [24].
Fosmid probes obtained from the BACPAC resources (CHORI) were combined to generate two specific probe contigs at the 2q31 HOXD locus. The centromeric probe contig contained the G248P89002G8, G248P81636C8, and G248P85016G11 fosmids and sized 75 Kb; the telomeric probe contig contained the G248P8191F1 and G248P8247G5 fosmids and sized 55 Kb. The centromeric probe contig overlapped the telomeric part of duplication A and the centromeric part of duplication B, while the telomeric probe contig hybridized the telomeric part of duplication B only. The centromeric and telomeric probe contigs were labeled by nick translation (Abbott Molecular Inc, IL, US) with Green- and Orange-dUTP, respectively (ENZO®, Life Sciences Inc, NY, USA). FISH experiments were performed with the two probe contigs following standard protocols.
Results
Cytogenetic and aCGH results
Standard karyotyping was normal in both families. In patient III-2 of family 1, we identified by aCGH two microduplications at 2q31 (hg19 chr2:g.(176879667_176880567)_(176987675_176988244)dup, (177002114_177002216)_(177092943_177093098)dup (Fig. 2 and Supplementary Fig. S1A; Decipher #363938). The 107 kb centromeric duplication (duplication A on Fig. 2 and Supplementary Fig. S2) included the HOXD10-13 and EVX2 genes, the 5′ half of the HOXD9 gene and the regulatory element Prox. The 90 kb telomeric duplication (duplication B on Fig. 2 and Supplementary Fig. S2) included the HOXD1, HOXD3, and HOXD4 genes. The 13.8 kb two copy-number chromosomal region located between the two microduplications contained the HOXD8 gene and the 3′ half of the HOXD9 gene. Since HOXD9 was only partially duplicated, it is likely that this allele was unable to encode any functional HOXD9 proteins. No other pathogenic genomic imbalances were identified in the patient. The same duplication was identified in the father II-1. qPCR confirmed the duplications in patients II-1 and III-2 of family 1 and the analyses in individuals I-1 and I-2 showed that the duplications occurred de novo in patient II-1. The analysis of both parents of patient II-1 by paired-end whole-genome sequencing did not show any balanced chromosomal rearrangement (such as an inversion or a more complex balanced rearrangement) that might have predisposed to the duplication identified in patient III-1.
HOXD locus with the location of the duplications identified in families 1 and 2. Massively parallel mate-pair sequencing, long-range PCR, and fiber FISH were used to characterize the genomic organization of the HOXD locus in both families. In family 1, the combination of the two probe contigs was designed to observe a 90 kb unlabeled genomic fragment located within the series of probe contigs. Fiber FISH analysis showed that the 90 kb unlabeled genomic fragment was detected at the telomeric end of the rearrangement (indicated by the white double arrows). These results showed that duplications in the two families were inserted between the HOXD gene cluster and the telomeric enhancers. TEL telomeric region, CEN centromeric region, P proximal regulatory element. The black double arrows indicate the breakpoints that have been sequenced
In patient II-1 of family 2, we identified by aCGH a microduplication at 2q31 (hg19 chr2:g. (176854470_176860309)_(177135095_177138646)dup (duplication C on Fig. 2 and Supplementary Fig. S1B; Decipher #363939). The 274 kb duplication included all HOXD genes, EVX2, the exons 1–4 of KIAA1715 (alias LNP for Lunapark), the regulatory element Prox and the 5′ end of MTX2. No other pathogenic genomic imbalances were identified in the patient. aCGH analyses showed that the father presented the duplication in a mosaic state. We estimated that ~20–30% of the lymphocytes carried the duplication.
In a previous study, Park et al. [25] identified a duplication including the HOXD gene cluster in four HAPMAP individuals of Asian origin (NA18542, NA18968, AK6, AK10). In contrast, we obtained normal results with our custom targeted 60 K array, excluding a duplication in the 2q31 region in the two individuals that we tested (NA18542, NA18968). Our results thus show that the duplications identified by Park et al. [25] in these two individuals were false positive results. We could not obtain DNA of the two other individuals (AK6, AK10). Therefore, duplication involving the HOXD gene cluster is not commonly observed in individuals of the general population (www.tcag.org).
Breakpoint sequencing
The orientation and the breakpoints of the duplicated fragments were determined by massively parallel mate-pair and paired-end sequencing and confirmed by long-range PCR and Sanger sequencing. In family 1, we were able to amplify and sequence the two breakpoints (Supplementary Fig. S2A, B). Alignment and mapping of mate-pair reads showed that the two duplicated fragments were in opposite orientations. In family 2, the rearrangement turned out to be more complex than anticipated with the duplication containing two fragments C′ and C″ (a 230 kb large centromeric duplication for fragment C′ and a 34 kb small telomeric duplication for fragment C″). A combination of mate-pair and paired-end whole-genome sequencing led us to identify the two breakpoints. Sequencing of the breakpoints revealed that the 230 kb large C′ centromeric fragment was inverted, followed by the 34 small C″ telomeric duplication. The C′ and C″ fragments were inserted between the wild-type HOXD gene cluster and the telomeric enhancers (Supplementary Fig. S2C, D). Since no fresh cells were available from family 2 to perform fiber FISH, we were not able to precise whether the 34 kb C″ telomeric fragment was in direct or inverted orientation. However, the orientation of the 34 kb C″ telomeric fragment does not modify the distance between the duplication containing HOXD13 and the telomeric enhancers.
Fiber FISH and genomic organization
Breakpoint sequencing led us to hypothesize two genomic organizations for the complex rearrangement identified in family 1. To further define these organizations, we performed fiber FISH analyses using a combination of two probe contigs (Supplementary Fig. S3A). This combination was designed to observe a 90 kb unlabeled genomic fragment located within the series of probe contigs (Supplementary Fig. S3B). Fiber FISH analysis showed that the 90 kb large unlabeled genomic fragment was detected at the telomeric end of the rearrangement (Fig. 2 and Supplementary Fig. S3B). This result allowed us to distinguish between the two possible genomic organizations and showed that duplications A and B in family 1 were inserted between the HOXD gene cluster and the telomeric enhancers. Also, duplication A was in an inverted orientation (Fig. 2 and Supplementary Fig. S3B).
Discussion
In humans, the HOXD gene cluster is found in a several megabase-sized mouse synthenic region, which expectedly contains all sequences identified as important for the regulation of HoxD genes during murine limb development. Like in mice, variants affecting function of HOXD genes lead to limb anomalies in humans. For example, loss-of-function variants in the HOXD13 gene lead to synpolydactyly and/or brachydactyly. Deletions of the entire HOXD cluster and a smaller deletion removing only the HOXD9–13 and EVX2 genes were reported to result in mild limb malformations, including fifth-finger clinodactyly, variable cutaneous syndactyly of the toes, hypoplastic middle phalanges of the feet and synpolydactyly [26, 27]. This phenotype is similar to the mild synpolydactyly/brachydactyly phenotype commonly found in patients with HOXD13 variants, indicating that deletions of the entire HOXD cluster do not necessarily produce major limb defects, in particular in proximal and intermediate segments, mostly due to the redundant functions of HOXA genes [28].
Several balanced chromosomal rearrangements, such as translocations or inversions, with breakpoints located in the vicinity of the HOXD cluster have been described in patients with various limb malformations (Fig. 3). In one patient, a symmetrical limb phenotype consisting of ulnar aplasia, radial shortening, absence of the third to fifth rays, and scoliosis was described in association with the balanced translocation t(2;10)(q31.1;q23.33) [29]. Humerus and lower limbs were normal. The breakpoint was localized ~950 kb away from the 3′ end of the HOXD cluster (Fig. 3; t1). A second patient carrying the balanced translocation t(2;8)(q31;p21) presented with mesomelic dysplasia of the upper limbs and vertebral defects [30]. No lower limbs anomalies were noticed. The translocation breakpoint was located 56 kb away from the 3′ end of the HOXD cluster (Fig. 3; t2) [31]. In a third patient, a pericentric inversion (inv(2)(p15q31)) was associated with bilateral aplasia of the fibula and the radius, bilateral hypoplasia of the ulna, unossified carpal bones, and hypoplasia and dislocation of both tibiae (Fig. 3; Inv) [29]. The balanced rearrangements in these patients likely separated the telomeric enhancers from the HOXD gene cluster leading to a predicted misregulation of these genes.
Overview of the HOXD region. The breakpoints of the balanced chromosomal rearrangements identified in patients with mesomelic dysplasia and in the mouse mutant Ulnaless are indicated in the upper part of the panel and the duplications in the lower part. Filled black rectangles indicate the genes; gray ovals indicate the regulatory elements. CEN centromeric region, TEL telomeric region, inv inversion [29], t1 and t2 translocation breakpoints [29,30,31], P proximal regulatory element, GCR global control region
Duplications of the HOXD cluster have been identified in rare patients. A 3.8 Mb large duplication at 2q31.1q31.2 comprising 27 genes including the entire HOXD cluster has been identified in a father and his daughter presenting with bilateral hand syndactyly and nystagmus [32]. A second patient with a 2q24.3q32.1 duplication presented with early-infantile-onset epilepsy, hypoplastic left heart syndrome, and global developmental delay [33]. None of these two patients presented mesomelic dysplasia. These large duplications include the whole synthenic mouse–human region containing the telomeric CNS 39 and 65. Thus, the proper regulation of HOXD genes was likely preserved explaining the absence of mesomelic dysplasia.
More interestingly, smaller duplications have been reported in patients with MDK (Fig. 2). A complex genomic imbalance composed of two microduplications at 2q31.1q31.2 encompassing respectively ~481 and 507 kb large DNA fragments separated by a segment of normal copy-number, were identified in the originally described Thai family with mesomelic dysplasia Kantaputra type [14, 19]. The centromeric duplication encompasses the entire HOXD cluster, as well as the neighboring genes EVX2, and MTX2, whereas the telomeric duplication includes HNRNPA3 and NFE2L2 (Fig. 3). It is noteworthy that no CNV within chromosome 2q or elsewhere in the genome was found in the three other families reported with MDK [16,17,18]. An ~1 Mb large duplication was identified in an unrelated family with patients affected by mesomelic shortening of the upper limbs and digital anomalies but only mild lower limb involvement [20]. In this case, the duplication included the HOXD cluster, EVX2, and MTX2. The phenotypes observed in this second family resemble MDK in terms of marked radius and ulna shortening as compared with relatively mild tibial or fibular shortening, although it lacked equinus deformity, calcaneofibular fusion, carpo-tarsal synostosis, or tibiofibular synostosis. In latter two families, the duplications included centromeric and telomeric enhancers. Importantly however, the orientations and organizations of the two duplications were not characterized by further molecular studies and thus remain unknown. In the families studied here, the upper limb involvement was strikingly similar to that observed in MDK, i.e., predominant on the ulnae instead of the radii like observed in dyschondrosteosis in which Madelung deformity may be present. The phenotype mainly differed from MDK by the absence of lower limb involvement, also resulting in the absence of short stature.
Strikingly, the mouse mutant ulnaless (Ul) shows a phenotype similar to the patients with MDK (i.e., a strong reduction of both forearms and forelegs). Ul is caused by an ~770 kb large balanced paracentric inversion, with one breakpoint centromeric to the HoxD cluster disrupting the Lnp gene and the other breakpoint telomeric to the HoxD cluster (Fig. 3) [8]. The inversion includes Evx2, the HoxD cluster, Mtx2, and CNS 39. Gene expression studies have shown that Ul embryos show an ectopic expression of Hoxd13 and Hoxd12 in the proximal limb (zeugopod) and a reduction of Hoxd13, Hoxd12, and Hoxd11 in the autopod [34, 35]. The skeletal reductions of the Ul mutant were interpreted as a consequence of posterior prevalence, whereby the proximal misexpression of Hoxd13 and Hoxd12 results in the transcriptional and/or functional inactivation of Hox group 11 and 10 genes via a dominant-negative effect.
These data combined with our present results suggest that our patients suffered from a misregulation of some HOXD genes during limb development. In this context, two molecular mechanisms can be envisaged as causative of the mesomelia. The first involves a loss-of-function of several genes critical for the development of the forearm, such as HOXD11 to HOXD9. In this view, the duplications would increase the distances between them and their telomeric enhancers, leading to a severe downregulation of their transcription (5; gray arrows). This possibility is supported by the pericentric inversion reported by Dlugaszewska B et al. [29], which separated HOXD genes from these enhancers and induced strong defects in all limb long bones. However, it is at odd with the deletion of the entire HOXD cluster, which does not elicit any substantial phenotype anywhere else than in digits [26, 27], due to the compensatory effect of HOXA genes [6].
Alternatively, these two distinct rearrangements could lead to the ectopic transcription of HOXD13 into the developing forearm territory. Since this protein was shown to act as a dominant-negative over other HOX proteins [36], its presence in these cells would abrogate the functions of all group 10 and 11 HOX proteins, thus inducing the observed mesomelia. Several lines of evidence support this latter interpretation, at the expense of the former. First, ectopic gains of function of Hoxd13 in forearm cells in mice were shown to induce severe forearm malformations [34, 35, 37], similar to those reported here. Secondly, the molecular mapping of these two duplications makes it likely that the telomeric enhancers increase their contacts with HOXD13 in the mutant configurations. In family 1, the inverted HOXD13 gene is now located much closer to these enhancers, right after HOXD4 in term of respective position (Fig. 4, dashed arrows). This situation is even accentuated in family 2, where inverted HOXD13 is now at the first position with respect to the 39 and 65 enhancers (Fig. 4, dashed arrows). In both conditions it is thus likely that HOXD13 became activated in the presumptive forearm early on, where it exerted its deleterious effect. Noteworthy, these two explanations are not exclusive and the observed mesomelia may derive from both a downregulation of HOXD9 to HOXD11 and a gain-of-function of HOXD13.
Model describing two alternative molecular mechanisms underlying the phenotypes described in the two families, both involving the misregulation of HOXD genes. In model 1, the duplicated DNA segments interfere with the proper regulation of HOXD9 to HOXD11, leading to their loss-of-function (gray arrows). This may result in an abnormal early phase of transcription of HOXD9 to HOXD11, leading to mesomelic dysplasia in the patients. In model 2, the forearm enhancers now take control of HOXD13, due to its ectopic position following the duplicated inversions (dashed arrows). Abnormal expression of HOXD13 in the developing forearm leads to severe malformations caused by the dominant-negative effect of this protein over other HOX proteins active in the limbs. The vertical double arrows indicate the breakpoints sequenced at the base-pair level
Recently, a fetus with mesomelia confined to the upper limbs (shortening and bowing of radius and ulna) and carrying two de novo 2q31.1 microdeletions has been reported [38]. The microdeletions did not include any HOXD genes but rather disrupted both the telomeric and centromeric TADs of the HOXD cluster. Given that the mesomelia is limited to the upper limbs and that the TAD telomeric to the HOXD cluster is responsible for regulation of more proximal limb morphogenesis, it is likely that the more distal 2q31.1 deletion is responsible for the mesomelia through a gain-of-function of HOXD13 in proximal limb [39]. The same explanatory framework can be applied to the mesomelic dysplasia Kantaputra type [14, 19], provided the duplication containing the HOXD cluster would be in an opposite orientation, which has not yet been clearly established. In such a case, HOXD13 would again be positioned at the vicinity of the forearm enhancers, leading to the same phenomenon as described above. In the families reported here, one copy of HOXD13 remains normally positioned close to its own centromeric enhancers, in agreement with the absence of any severe phenotype in the digits of these patients.
References
Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201.
Dollé P, Izpisúa-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–72.
Kmita M, Duboule D. Organizing axes in time and space; 25 years of collinear tinkering. Science. 2003;301:331–3.
Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–45.
Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10:93–103.
Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–66.
Gonzalez F, Duboule D, Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev Biol. 2007;306:847–59.
Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–17.
Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80.
Lonfat N, Duboule D. Structure, function and evolution of topologically associating domains (TADs) at HOX loci. FEBS Lett 2015;589:2869–76.
Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, Le Merrer M, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–68.
Reardon W, Hall CM, Slaney S, Huson SM, Connell J, al-Hilaly N, et al. Mesomelic limb shortness: a previously unreported autosomal recessive type. Am J Med Genet. 1993;47:788–92.
Kantaputra PN, Gorlin RJ, Langer LO Jr. Dominant mesomelic dysplasia, ankle, carpal, and tarsal synostosis type: a new autosomal dominant bone disorder. Am J Med Genet. 1992;44:730–7.
Kantaputra PN. Thirteen-year-follow up report on mesomelic dysplasia, Kantaputra type (MDK), and comments on the paper of the second reported family of MDK by Shears et al. Am J Med Genet A. 2004;128A:1–5.
Kwee ML, van de Sluijs JA, van Vugt JM, Wijnaendts LC, Gille JJ. Mesomelic dysplasia, Kantaputra type: clinical report, prenatal diagnosis, no evidence for SHOX deletion/mutation. Am J Med Genet A. 2004;128A:404–9.
Shears DJ, Offiah A, Rutland P, Sirimanna T, Bitner-Glindzicz M, Hall C. Kantaputra mesomelic dysplasia: a second reported family. Am J Med Genet A. 2004;128A:6–11.
Siwicka KA, Kitoh H, Nishiyama M, Ishiguro N. A case of mesomelic dysplasia Kantaputra type–new findings and a new diagnostic approach. J Pediatr Orthop B. 2008;17:271–6.
Kantaputra PN, Klopocki E, Hennig BP, Praphanphoj V, Le Caignec C, Isidor B, et al. Mesomelic dysplasia Kantaputra type is associated with duplications of the HOXD locus on chromosome 2q. Eur J Hum Genet. 2010;18:1310–4.
Cho TJ, Kim OH, Choi IH, Nishimura G, Superti-Furga A, Kim KS, et al. A dominant mesomelic dysplasia associated with a 1.0-Mb microduplication of HOXD gene cluster at 2q31.1. J Med Genet. 2010;47:638–9.
Fryns JP, Hofkens G, Fabry G, van den Berghe H. Isolated mesomelic shortening of the forearm in father and daughter: a new entity in the group of mesomelic dysplasias. Clin Genet. 1988;33:57–9.
Mégarbané A, Ghanem I. Severe autosomal dominant upper-limb mesomelic dysplasia: report of a second family. Clin Genet. 2005;68:567–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods. 2001;25:402–8.
Heiskanen M, Kallioniemi O, Palotie A. Fiber-FISH: experiences and a refined protocol. Genet Anal. 1996;12:179–84.
Park H, Kim JI, Ju YS, Gokcumen O, Mills RE, Kim S, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet. 2010;42:400–5.
Prieur M, Lapierre J, Le Lorch M, Ozilou C, Amiel J, Sanlaville D, et al. HOXD gene cluster haploinsufficiency does not generate gross limb abnormalities. Eur J Hum Genet. 2000;8:74.
Slavotinek A, Schwarz C, Getty JF, Stecko O, Goodman F, Kingston H. Two cases with interstitial deletions of chromosome 2 and sex reversal in one. Am J Med Genet. 1999;86:75–81.
Kmita M, Tarchini B, Zàkàny J, Logan M, Tabin CJ, Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435:1113–6.
Dlugaszewska B, Silahtaroglu A, Menzel C, Kübart S, Cohen M, Mundlos S, et al. Breakpoints around the HOXD cluster result in various limb malformations. J Med Genet. 2006;43:111–8.
Ventruto V, Pisciotta R, Renda S, Festa B, Rinaldi MM, Stabile M, et al. Multiple skeletal familial abnormalities associated with balanced reciprocal translocation 2;8(q32; p13). Am J Med Genet. 1983;16:589–94.
Spitz F, Montavon T, Monso-Hinard C, Morris M, Ventruto ML, Antonarakis S, et al. A t(2;8) balanced translocation with breakpoints near the human HOXD complex causes mesomelic dysplasia and vertebral defects. Genomics. 2002;79:493–8.
Ghoumid J, Andrieux J, Sablonnière B, Odent S, Philippe N, Zanlonghi X, et al. Duplication at chromosome 2q31.1-q31.2 in a family presenting syndactyly and nystagmus. Eur J Hum Genet. 2011;19:1198–201.
Lim BC, Min BJ, Park WY, Oh SK, Woo MJ, Choi JS, et al. A unique phenotype of 2q24.3-2q32.1 duplication: early infantile epileptic encephalopathy without mesomelic dysplasia. J Child Neurol. 2014;29:260–4.
Hérault Y, Fraudeau N, Zákány J, Duboule D. Ulnaless (Ul), a regulatory mutation inducing both loss-of-function and gain-of-function of posterior Hoxd genes. Development. 1997;124:3493–500.
Peichel CL, Prabhakaran B, Vogt TF. The mouse Ulnaless mutation deregulates posterior HoxD gene expression and alters appendicular patterning. Development. 1997;124:3481–92.
Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–64.
van der Hoeven F, Zákány J, Duboule D. Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell. 1996;85:1025–35.
Peron A, Boito S, Rizzuti T, Borzani I, Baccarin M, Bedeschi MF, et al. Prenatal upper-limb mesomelia and 2q31.1 microdeletions affecting the regulatory genome. Genet Med. 2018;20:1483–4.
Kragesteen BK, Duboule D, Mundlos S, Spielmann M. Response to Peron et al. Genet Med. 2018;20:1481–2.
Acknowledgements
We are grateful to the patients and their families who participated in this study. We are most grateful to the Genomics platform of Nantes (Biogenouest Genomics) core facility for its technical support. DNA panels from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds) were used in this study.
Author information
Authors and Affiliations
Contributions
Authors’ roles: Study design: CLC, OP, and AT. Study conduct: CLC, OP, and AT. BdC, CB, AM, and AT clinically characterized the patients and collected blood samples. Array CGH analysis and interpretation of the data: CLC, AB, OP, and MSC. Whole-genome sequencing and data interpretation: OP, PL, RR, CSB, and DS. Fiber FISH experiments and data interpretation: OP, MSC. Drafting manuscript: CLC, OP, DD, and AT. Revising manuscript content: CLC, OP, DD, MLV, AM, and AT. Approving final version of manuscript: CLC, OP, AB, BdC, CB, DD, MLV, PL, RR, CSB, MSC, DS, DD, AM, and AT. CLC and AT take responsibility for the integrity of the data analysis.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Le Caignec, C., Pichon, O., Briand, A. et al. Fryns type mesomelic dysplasia of the upper limbs caused by inverted duplications of the HOXD gene cluster. Eur J Hum Genet 28, 324–332 (2020). https://doi.org/10.1038/s41431-019-0522-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-019-0522-2
This article is cited by
-
Inhibition of HOXD11 promotes cartilage degradation and induces osteoarthritis development
Journal of Orthopaedic Surgery and Research (2024)
-
Mesomelic dysplasias associated with the HOXD locus are caused by regulatory reallocations
Nature Communications (2021)