Abstract
In the present study, 87 unrelated individuals from the Marquesas Archipelago in French Polynesia were typed using mtDNA, Y-chromosome and autosomal (STRs) markers and compared to key target populations from Island South East Asia (ISEA), Taiwan, and West and East Polynesia to investigate their genetic relationships. The Marquesas, located at the eastern-most fringes of the Austronesian expansion, offer a unique opportunity to examine the effects of a protracted population expansion wave on population structure. We explore the contribution of Melanesian, Asian and European heritage to the Marquesan islands of Nuku-Hiva, Hiva-Oa and Tahuata. Overall, the Marquesas Islands are genetically homogeneous. In the Marquesan Archipelago all of the mtDNA haplogroups are of Austronesian origin belonging to the B4a1 subhaplogroup as the region marks the end of a west to east decreasing cline of Melanesian mtDNA starting with the West Polynesian population of Tonga. Genetic discrepancies are less pronounced between the Marquesan and Society islands, and among the Marquesan islands. Interestingly, a number of Melanesian, Polynesian and European Y-chromosome haplogroups exhibit very different distribution between the Marquesan islands of Nuku Hiva and Hiva Oa, likely resulting from drift, differential migration involving various source populations and/or unique trading routes.
Similar content being viewed by others
Introduction
The Austronesian Expansion involved two thirds of the circumference of the world from Madagascar in East Africa to Easter Island (Rapa Nui) 2300 miles off the western coast of Chile. Recent archeological evidence suggests a relative recent colonization of the Marquesas, approximately 1190–1290 AD [1]. Subsequently, the Spanish explorer Álvaro de Mendaña reached the Marquesas on July 21, 1595 and named them after his sponsor the Marquis of Cañete. Since 1842 France has claimed the islands.
Two main models have been proposed to explain the genesis of the Austronesian spread. In one, the Out of Taiwan model, the island of Formosa was populated by proto-Austronesian-speaking groups from the mainland about 12,000–8000 years ago (ya), after which they initiated their exodus into Oceania and the Indian Ocean [2]. Linguistics corroborates this scenario, since Taiwan exhibits the highest diversity of Austronesian languages in the world [3, 4]. Furthermore, some elements of the Polynesian material culture, such as the Lapita pottery tradition, have been traced to the red-slip pottery made by the Austronesians of Taiwan about 8000 ya [5]. Also, whole genome DNA data of multiple Lapita-related skeletal remains from Vanuatu and Tonga dated at 3100–2300 ya suggest that the Lapita people migrated from South East Asia (SEA) via Taiwan and the Philippines to Polynesia [6]. Ni-Vanuatu ancient DNA also indicate that subsequent to the initial Austronesian expansion from SEA, at least two additional streams of migrations introduced Papuan DNA to Polynesia posing the possibility that the Melanesian component in remote Oceania derives from different sources [6] including the Bismarck Archipelago [7, 8]. In the alternative scenario, known as the SEA origin model, Taiwan is seen as a side branch of the expansion that did not progress past the island [9, 10]. The two theories are not mutually exclusive as both postulate Neolithic cultures, such as the Hemudu, Liangzhu or Daic-speakers, traveling from continental SEA to ISEA and beyond into Oceania [11, 12].
It is likely that Austronesian farmers migrated south from Taiwan into the Philippine Archipelago. The current inhabitants of the Philippines speak the Malayo-Polynesian languages associated with the Austronesian expansion and Polynesia [13]. During the next few thousand years, Austronesians moved in a southeasterly direction populating the islands of the Celebes Sea, Borneo and then Sulawesi in ISEA. This route delineates an insular arc that includes the islands of Melanesia and Micronesia, which according to the archeological record were populated approximately 5500 ya and 1500 ya, respectively. Subsequently, Austronesians traveled along the northern coast of New Guinea and then migrated in an easterly direction progressively reaching the islands of the Bismarck, Solomon, Santa Cruz and the Vanuatu Archipelagos [14]. Fiji, was the first uninhabited island that Austronesians encountered around 3500 ya before moving into the unknown vastness of the Pacific Ocean. The Tonga Archipelago, just east of Fiji, in Western Polynesia, was populated around 3300 ya and the Samoan Islands approximately 300 years later [15,16,17]. A recent proposed late chronology based on 1434 reliable radiocarbon dates related to the settlement of East Polynesia indicates that Austronesians reached the Society Islands about 1020 ya [1, 18]. From the Society Islands, the Marquesas (around 830–730 ya) Rapa Nui (820 ya), Hawaii (800–850 ya) and New Zealand (740 ya) were settled [1, 19].
This trajectory and time line certainly allowed for interactions between Austronesian populations and the indigenous Papuan groups as they traveled through Melanesia eastward into Oceania. The presence of the Lapita culture and the Austronesian language in coastal areas and northern islands of Melanesia is testimony to the artistic and linguistic transfer that took place. Furthermore, utilitarian items such as pots made in the Rewa Delta region of Fiji have been found in Samoa and even in the Marquesan islands [20] while basalt adzes and other lithic tools from the Marquesas were traded 1425 kilometers southwest to the Society Islands, 1750 kilometers southeast to Mangareva [21] and some 2400 kilometers north west to the Phoenix and Line Archipelagoes [22]. Additionally, Marquesan folklore speaks of contacts with Rarotonga, 2600 kilometers southwest in the southern Cook Islands, to trade for prized bird feathers [23]. These archeological findings and shared cultural traditions [24, 25] point to regular contacts among the Pacific islands, including the Marquesas.
Genetic signals of Austronesian-Melanesian bidirectional interactions have been detected in the genome-wide and uniparental DNAs of Melanesian and Polynesian populations. Genome-wide DNA markers indicate that among specific coastal and insular populations within the Melanesian domain there is a small Asian component [26]. This whole-genome Austronesian genetic signature is never higher than 20% and is observed in less than 50% of the islands that speak Austronesian languages and never observed in Papuan-speaking populations. The highest frequencies of whole-genome Austronesian DNA are seen in northern-island Melanesian populations [6]. In these northern Melanesian islands, the uniparental Austronesian signals tend to be stronger with 29.4–72.5% mtDNA and 5.3–37.7% Y-chromosome Austronesian DNA [27]. The dichotomy exhibited between the contributions of Austronesian mtDNA and Y-chromosome DNA has been attributed to the patrilocal systems of the original Melanesian populations [27]. The restricted Austronesian genetic imprint in most of Melanesia suggests that Austronesians had a limited genetic impact on the autochthonous Papuan groups and provides a clue to the relative short stay and temporary settlements of Austronesians as they continued dispersing into Oceania.
In the opposite direction, gene flow from Papuan populations to Austronesian biparental DNA tends to be more robust. Using autosomal STR loci about 24% and 76% of the Samoan and 35% and 65% of the Tongan autosomal gene pools are of Melanesia and SEA descent, respectively [28]. On the other hand, uniparental markers provide different views, depending on whether Y-chromosome or mtDNA markers are used. Employing mtDNA, Asian ancestry is 93.8% and Melanesian 6% among Polynesian populations while 65.8% and 28.3% of Y chromosomes are Melanesian and Asian, respectively [27]. The bias in favor of Asian mtDNA and Melanesian Y-chromosome types in Polynesia is likely the result of the matrilocal family customs of Austronesian society [27, 29] or male-mediated migration of Papuan-ancestry into Remote Oceania mainly mating with local women carrying Austronesian ancestry.
To shed light on some of the issues presented above and the population dynamics of the Austronesian expansion, we report for the first time on the mtDNA, Y-chromosome and autosomal constitution of the Marquesas Archipelago located at the eastern-most extreme of the Polynesian domain.
Materials and methods
Populations, sample collection, and DNA isolation
Buccal swabs were collected from a total of 87 unrelated individual from the Marquesas Archipelago in French Polynesia. The populations of the three islands sampled included 51 males from Nuku-Hiva, 28 from Hiva-Oa and 8 from Tahuata. Genealogical information was collected for a minimum of three generations. DNA extraction was performed using the standard phenol:chloroform procedure [30]. NanoDrop 1000 Spectrophotometer (Thermo Scientific) was used for DNA quantitation. Samples were stored as stock solutions in 10 mM Tris-EDTA at −80 °C. All samples were procured from donors voluntarily while closely adhering to the ethical guidelines stipulated by Colorado College, Colorado Springs, Colorado USA. All donors gave their informed consent prior to inclusion in the study, following the ethical principles and guidelines of the Declaration of Helsinki for the protection of human subjects. The IRB of Colorado College approved this study.
Seven and 15 reference populations were employed for comparison across the Y-chromosome SNP and autosomal STR markers examined, respectively. The geographical locations, abbreviations used to define populations throughout the article, number of individuals and references are provided in Supplementary Table 1.
STR genotyping
Twenty-one autosomal STR loci (CSF1PO, D5S818, D7S820, D21S11, D2S441, D1S1656, TH01, D16S539, D3S1358, D18S51, D2S1338, TPOX, vWA, D8S1179, D19S433, D12S391, SE33, D13S317, FGA, D22S1045, and D10S1248) were analyzed with the I-DNASE21 amplification system [31] as previously described [2, 23].
Accession numbers and URLs
All mtDNA sequences have been deposited into EMPOP at http://www.empop.org under accession number EMP00715 after QC process. In addition, sequences are available online in GenBank at https://www.ncbi.nlm.nih.gov/ under accession numbers MG866082-MG866168. The GenBank accession number of the rCRS mtDNA sequence is NC_012920.1. The Y-chromosome data have been successfully submitted and are now included in the YHRD database at https://yhrd.org/ under the following accession numbers: Nuku-Hiva YA004253; Hiva-Oa YA004254.
Statistical analyses
Allelic frequencies were calculated utilizing GenePop [32] as previously described [33]. Population sub-structuring was explored using the Structure software v.2.3.3 with the admixture model at k = 2–20 [34]. The k value exhibiting the highest degree of structure was calculated according to Evanno et al. (2005) [35]. Average gene diversity over loci [36] and Slatkin’s linearized Rst values [37] were calculated using the ARLEQUIN 3.5 statistical package. Significance was assessed at p = 0.05. Based on Rst values, a multidimensional scaling (MDS) analysis was performed using PROXSCAL [38].
Full mitochondrial control regions (16,024–576 bp) were amplified, sequenced and interpreted as reported in [39] using rCRS as a reference sequence [40] and ISFG guidelines [41, 42]. Y chromosome lineages were ascertained through the analysis of 19 haplogroup-defining Y-SNPs as already described [43]. Primers used for HRM analysis and sequencing are shown in Supplementary Table 2. A correspondence analysis (CA) analysis was performed based on the Y-SNP haplogroup frequencies using the PAST v3.15 program [44]. For both mitochondrial and Y-chromosome data, genetic distances (FST) were calculated and Bonferroni corrections were performed as formerly reported [45]. Test Statistic (ts) was used to assess significance of percentages [46].
Results
Autosomal STRs
The autosomal STR genotypes are provided in Supplementary Table 3. None of the men exhibit parentage relationship. The autosomal STR genotypes were utilized to generate Rst genetic distances and the corresponding significant values at p = 0.05 (Supplementary Table 4). For this analysis, due to the limited number of donors from Tahuata, this group was added to the geographically-nearby population of Hiva Oa. A number of pairs of populations exhibit statistically non-significant differences, including CEB-AMI, CEB-PUY, AMI-PUY, ATA-SAI, PAW-PUY, PAW-RUK, PUY-RUK, RAI-BOR, RAI-NUH, RAI-HIO, BOR-NUH, and BOR-HIO.
Using the Rst values a MDS plot was constructed (Fig. 1). In the resulting graph, there is a complete genetic divide between Polynesian populations and aboriginal Taiwanese/Cebú groups. The population from Madagascar segregates as a outlier in the top left quadrant while the island populations of the Society Archipelago (RAI and BOR) and Marquesas Archipelago (NUH and HIO) cluster close to each other between the upper and lower left quadrants. Tonga partitions just to the upper left of the Society and Marquesan populations. Overall, the Taiwanese aboriginal populations exhibit greater genetic heterogeneity compared to the Polynesian groups, although specific Formosan tribes (PAW, PUK, and RUK) plot close to each other.
The average genetic diversity over loci is highest in MAD (0.80702) (Supplementary Table 5). The next two highest diversity levels (0.78476 and 0.78436) belong to CEB and TON, respectively. The lowest diversity observed is 0.70402 in the YAM. Taiwanese and Eastern Polynesia populations possess intermediate values.
Sub-structuring was observed among all populations examined at all k values (Fig. 2). A general signature pattern is shared among all Polynesian populations (Tonga, Raiatea, Bora Bora, Nuku Hiva and Hiva Oa) at k = 4 through K = 7. Yet, starting at k = 4 the Tonga population from West Polynesia differentiates away from the East Polynesian groups of Raiatea, Bora Bora, Nuku Hiva and Hiva Oa. Although Tonga exhibits a similar set of components in all k values, its degree of genetic heterogeneity is clearly greater compared to the Eastern Polynesian groups from the Marquesas and Society Archipelagos. For example, the dark blue component and other color signals observed in Tonga at k = 5 and 7 are reduced considerably in the Eastern Polynesian populations while the purple element in the later archipelagos augment and predominate. Further differentiation between Western and Eastern Polynesian groups is seen at k = 8 as a burgundy component is seen in Tonga but not in the four Eastern Polynesian populations. Except for what seems to be a European component seen in Nuku Hiva in blue (k = 3), red (k = 4), green (k = 5), purple (k = 6), red and light blue (k = 7), and dark blue and purple (k = 8), no differentiation was observed among the island populations of Eastern Polynesia. Parallelisms in composition of components are evident at k = 2–4 among Taiwanese aboriginal tribes and the Philippine island of Cebú. Among the Taiwanese tribes at k = 4–10 a number of unique combinations of components are seen in different sets of aboriginal groups. For example, the Yami population from the small Orchid Island off the southeast coast of Taiwan, exhibits a singular profile dominated by a single component in k = 4–10. Also, the Ami shares a similar complex profiles with the southern Formosan populations of Paiwan, Puyuma and Rukai, as well as with the Philippine group from Cebú at k = 5–10. The northern central Atayal aboriginal population shares a prominent light green element at k = 7 and a light blue at k = 8 component with the central (Bunun) and northern (Saisiyat) mountainous region tribes. At k = 9 similar profiles are evident among Atayal, Bunun and Tsou, yet at k = 10 the central mountainous Tsou population exhibits a unique prominent orange component not seen in any other Taiwanese aboriginal group.
Mitochondrial DNA
The mtDNA control region from 16024 to 576 bp for Nuku-Hiva, Hiva-Oa and Tahuata is provided in Supplementary Table 6. Molecular statistics of these populations are provided in Supplementary Table 7. Overall, 15 polymorphic sites were found, which defined 21 different haplotypes representing a genetic diversity of 0.8311 ± 0.0237. Individually, the Tahuata population exhibits the lowest genetic diversity with 0.6071 ± 0.1640 relative to Nuku Hiva (0.8196 ± 0.0310) and Hiva Oa (0.8413 ± 0.0535).
The most common haplotype found in these populations is shared by 26 individuals (30 %), 16 from Nuku Hiva, five from Hiva Oa and five from Tahuata, presenting the following polymorphisms: 16182C, 16183C, 16189C, 16217C, 16247G, 16261T, 16519C, 73G, 146C, 263G, 315.1C, 523DEL, 524DEL (HGVS-nomenclature: m.16182A>C, m.16183A>C, m.16189T>C, m.16217T>C, m.16247A>G, m.16261C>T, m.16519T>>C, m.73A>G, m.146T>C, m.263A>G, m.316dupC, m.523_524delAC). Two additional haplotypes are frequently shared in these islands. One is exhibited by 18 individuals (20.7%), 10 from Hiva Oa and eight from Nuku Hiva and is defined by polymorphisms 16182C, 16183C, 16189C, 16217C, 16247G, 16261T, 16519C, 73G, 146C, 151T, 263G, 315.1C, 523DEL and 524DEL (HGVS-nomenclature: m.16182A>C, m.16183A>C, m.16189T>C, m.16217T>C, m.16247A>G, m.16261C>>T, m.16519T>C, m.73A>G, m.146T>C, m.151C>T, m.263A>G, m.316dupC, m.523_524delAC). The second haplotype is shared by 17 individuals (19.5%), 12 from Nuku Hiva, three from Hiva Oa and two from Tahuata, with polymorphisms 16126C, 16182C, 16183C, 16189C, 16217C, 16247G, 16261T, 16519C, 73G, 146C, 263G, 309DEL, 315.1C, 523DEL and 524DEL (HGVS-nomenclature: m.16126T>C, m.16182A>C, m.16183A>C, m.16189T>C, m.16217T>C, m.16247A>G, m.16261C>T, m.16519T>C, m.73A>G, m.146>T>C, m.263A>G, m.309delC, m.316dupC, m.523_524delAC).
The control mtDNA sequences of the three Marquesan island populations were compared and analyzed by FST distances (Supplementary Table 8). The results show no significant differences (p > 0.008) between the analyzed islands. Phylogenetically, all mtDNA haplotypes belong to the B4a1 sub-haplogroup. Most of the haplotypes can be classified into the B4a1a1 sub-haplogroup (89.7%), or one of its sub-branches, such as B4a1a1h (3.5%), B4a1a1k (1.1%) and B4a1a1a14 (1.1%). One sample belonging to the B4a1c2 sub-haplogroup was identified in the Nuku Hiva population.
Y-SNP analysis
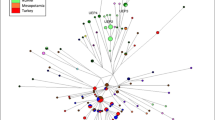
Y-SNP haplotypes and haplogroups (Supplementary Table 9) and their frequencies are shown in Fig. 3. Eight major haplogroups (C, O, K, E, G, I, Q, and R) were detected among the 87 individuals, haplogroup C-M130 being the most abundant, ranging from 39% in Hiva Oa to 75% in Tahuata (ts = 1.844, p = 0.0652). The observed levels of this Melanesian lineage are in accordance with those found in other regions of Polynesia [2, 47]. However, the high frequency of C-M130 observed in Tahuata could be the result of founder effects, bottlenecks or a sampling effect due to the reduced sample size.
The O-M175 (xM324, xP164) haplogroup is found at frequencies ranging from 2% to 12% and is further subdivided into sub-lineages O3a (M324) and O3a2c (P164). O3a (M324), typically found in South East Asia, East Asia and Austronesian regions [2, 27], was detected with frequencies between 2% and 12% in the three islands, while O3a2c-P164 was only observed in Nuku Hiva (12%). O3a2c-P164 has been observed in other Polynesian areas [2, 48, 49]. As for the other remaining haplogroups, a moderately high frequency of the Melanesian K-M9 marker (29%) is evident in Hiva Oa but was not detected in the other two islands. Conversely, European haplogroup I-M258 is only observed in Nuku Hiva (14%). Haplogroups Q, G, R and E are also observed in low frequencies (4–10%) throughout the islands.
Pairwise comparisons of the three Marquesas populations using FST genetic distance analysis based on Y-SNP haplotypes (Supplementary Table 10) indicated no significant differences (p > 0.008) within the archipelago. Therefore, as for mtDNA, no significant genetic substructure was detected based on the Y-SNPs in the Marquesas.
The PC (Fig. 4) plot exhibits a clear divide between the populations from West (Samoa and Tonga) and East (Marquesas and Society) Polynesia along the Y-axis. The position of the population from Hiva Oa in the Southwest of the Marquesas Archipelago as an outlier may result from the presence of a number of European-derive haplogroups in the population. Tonga segregates in the top left quadrant some distance away from the Samoan populations. Nuku Hiva and Tahuata appear at the periphery of the East Polynesian populations, distinctly away from the West Polynesian groups.
Based on the abundance of Asian and Melanesian Y chromosomes, the contributions of the above geographical regions to the Islands of the Marquesas and other Polynesian populations were assessed (Supplementary Table 11). European Y haplogroups are found only in the Marquesas and one Tongan individual had a Y chromosome of African descent. However, since European and African Y chromosomes are likely to derive from recent admixture, they were excluded from these calculations. Although frequency differences were observed among the Marquesan, Society, Samoa and Tonga archipelagos, statistical analyses based on pair wise comparisons of Asian and Melanesian Y chromosomes only indicate significant differences between the Marquesas and Tonga (Marquesas-Tonga Chi 9.9176, p = 0.001637; Marquesas-Samoa Chi 0.21204, p = 0.64517; Marquesas-Society Chi 0.22506, p = 0.63521). The frequencies of Asian-derived Y chromosomes in the Societies, Marquesas, Samoan and Tongan Islands are 23.6%, 27.0%, 30.2% and 54.9%, respectively. Only pair wise comparisons involving Tonga with the Marquesas, Societies and the Samoan Archipelgo are statistical significant (ts = 3.166, p = 0.0016; ts = 3.575, p = 0.0004; ts = 2.965, p = 0.0030, repectively). The Samoan islands exhibit Asian frequencies that range from 26.2% (West Samoa) to 41.7% (Tutuila) (ts = 1.367, p = 0.1717). The Melanesian component is highest in the Societies (76.4%) followed by the Marquesas at 73.0% and Samoans at 69.8% (Societies-Marquesas ts = 0.473, p = 0.6365; Societies-Samoa ts = 0.976, p = 0.329; Samoa-Marquesas ts = 0.468, p = 0.6401). Tonga possesses the lowest prevalence of Melanesian Y chromosomes (45.1%) and exhibits statistically significant differences with the Societies (ts = 3.575, p = 0.0004), Samoans (ts = 3.091, p = 0.0030) and Marquesas (ts = 3.166, p = 0.0016) (Supplementary Table 11).
Discussion
The Marquesan Archipelago, located at the fringes of the Austronesian expansion range, offers a unique opportunity to study the population genetic dynamics of a dispersion wave at its pinnacle of activity just before the Polynesian long-range voyages of discovery ceased. Examination of the Rst values based on autosomal STR data indicates that the populations of the Marquesas and Societies are closely related. Five combinations of islands involving these two archipelagos exhibit non-significant genetic differences, with the Raiatea population in three of the pairs and Bora Bora in the other two pairs. In addition to a likely ancestor-descendent relationship between these two archipelagos, trade between them may also account for continuous gene flow among these islands. These two archipelagos are about 1500 kilometers apart.
In contrast, the statistically significant genetic differences between the populations of West and East Polynesia suggest less gene flow between the two sets of populations. This observation is also reflected in the MDS plot, where the Tonga population is seen segregating away from the East Polynesian groups possibly resulting from additional genetic inputs into Tonga after the divergence from Eastern Polynesian populations or genetic drift. The greater proximity of Tonga to Raiatea in the plot as compared to the other East Polynesian islands may reflect a closer genetic affinity between the two due to the critical role played by Raiatea as a commercial hub in Oceania. Genome-wide similarities between ancient Raiateans and modern-day Tongans are compatible with this divide [8]. Oral tradition indicates that voyages of discovery and trade were initiated in the Marae of Taputapuatea (major ceremonial site) in Raiatea as the center of the cult of the god ‘Oro and the core of a long-range voyaging network that extended in all directions within Polynesia [50]. The relatively high average gene diversity (the highest among the East Polynesian populations examined in this study) exhibited by the population of Raiatea is congruent with the unique role that this island played in trade and communication.
The Structure analyses based on autosomal STRs also corroborate a close affinity among the Eastern Polynesian Island populations. At k = 3 a clear divide is first observed between the West Polynesian population of Tonga and the East Polynesian groups, which is accentuated as the k values increase. Except for minor signals (e.g., red and light blue at k = 7) in Nuku Hiva, no apparent difference in genetic components is seen among the populations from the Marquesan and Society Archipelagos. These results substantiate the lack of significant genetic differences among the islands of these two archipelagos based on Rst values. In these Structure analyses, Tonga exhibits similar signals compared to the populations of the Marquesas and Society islands, yet the diversity of components is greater in Tonga. Cebú, as well as specific Taiwanese aboriginal groups exhibit a dark blue component at k = 5 and k = 6 that is shared with Polynesian populations. Among Polynesians this element is more evident in Tonga and is diluted in East Polynesia. The observed parallelisms in the Structure components may reflect an ancestral-descendent relationship stemming from Tongan migrants and/or trade involving East and West Polynesia. Furthermore, the limited genetic heterogeneity seen in the Marquesas and Society islands is likely the result of bottleneck events and/or random drift.
Of note, similarities were observed in the Structure analyses between the Ami Taiwanese aborigines and the natives of the Central Philippine province of Cebú. The general parallelisms in the component profiles are seen at k = 3 through k = 10 and extend, to a lesser extent, to the Paiwan, Puyuma and Rukai tribes of southern Taiwan. This broad resemblance in Structure profiles involving the Amis (inhabitants of the eastern coastal plains of Formosa), the tribes of southern Taiwan and the Cebú population of Central Philippines are compatible with the Out of Taiwan/ISEA model for the Austronesia dispersal since the similarities delineate a geographical arc from Formosa to Oceania.
The mtDNA data from the Marquesan islands indicate that all the haplogroups are of Asian origin; no Melanesian mtDNA signal was detected in the Marquesas. In these islands, at the fringe of the Austronesian expansion, the preponderance of Asian maternal lineages reached an apex. In the Marquesas all individuals belong to haplogroup B4a1, and 89.7% of them are B4a1a1. Minimal frequencies of B4a1a1h (3.5%), B4a1a1k (1.1%), B4a1c2 (1.1%), and B4a1a1a14 (1.1%) were detected as well. This homogeneity may be due to bottleneck episodes, drift and/or founder effect events. Overall in Polynesia, the proportion of Melanesian mtDNA haplogroups is about 6.0%, and the rest are basically Austronesian haplogroups [46]. Specifically, in Tonga and Samoa, Melanesian mtDNA has been assessed to be 7.7% (haplogroups M-28 and P1) and 6.0% (haplogroup Q1), respectively. In the Society Archipelago in East Polynesia only approximately 3.7% (Q1f1, 2.47% and Q2a5, 1.23%) of the mtDNA is of Melanesian origin (Societies-Marquesas ts = 2.430, p = 0.0151) [18]. These values suggest a divide between the Society and Marquesas Archipelagos due to drift and/or bottleneck events during the step-wise expansion.
The genetic heterogeneity observed in the Y chromosomes of the Marquesas is greater than that of mtDNA types. Distinct splits of Asian and Melanesian Y chromosomal haplogroups are observed in the Marquesas (27% and 73%, respectively), Society (24% and 76%, respectively), Samoan, (30% and 70%, respectively) and Tonga (55% and 45%, respectively) islands. The Marquesas and Societies exhibit similar levels of Asian and Melanesian Y types (ts = 0.473, p = 0.6365). Also, the Marquesas contain 14.9% of European Y chromosomes (17.6% in Nuku Hiva and 14.3% in Hiva Oa), not seen in Societies [18]. This European element in the Marquesas is likely reflected in the blue (k = 3), red (k = 4), green (k = 5), purple (k = 6), red and light blue (k = 7), and dark blue and purple (k = 8) components of Nuku Hiva in the Structure analyses.
A number of Y haplogroups exhibit very different distribution between the islands of Nuku Hiva and Hiva Oa. This phenomenon is seen in both Melanesian and Asian Y chromosome types. For example, haplogroup C is found at 56.9% (n = 29/51) in Nuku Hiva and only 39.3% (n = 11/28) in Hiva Oa (ts = 1.505, p = 0.1322) while haplogroup K is observed at 28.6% (n = 8/28) in Hiva Oa, but absent in Nuku Hiva (ts = 4.798, p = 0.000002) and O3a2c is 11.8% (n = 6/51) in Nuku Hiva, but absent in Hiva Oa (ts = 2.981, p = 0.0029). Although these dichotomies may simply reflect sampling errors, random drift, or founder effects as the islands were colonized, it is possible that the differences may result from differential colonization from unique source populations and/or variation in trading routes among the Marquesan islands.
Conclusion
Here we investigate the uniparental and genome-wide composition of three islands of the Marquesas Archipelago and their phylogenetic relationships to other Polynesian and Melanesian populations of the Pacific. The overall picture presented by the Marquesas is one of relative genetic homogeneity, but not of isolation. The lack of genetic diversity in the Marquesas is clearly evident in the mtDNA types where all the haplogroups are of Austronesian origin belonging to the B4a1 sub-haplogroup, most of them falling into the B4a1a1 lineage. On the other hand, the nearby Society Archipelago exhibits 3.7% Melanesian mtDNA. It was observed that the Marquesas mark the end of a trend of west to east diminution of Melanesian mtDNA starting with the West Polynesian population of Tonga. Genetic differences in the proportion of Y chromosome-specific haplogroups are also seen between Western (Tonga and Samoa) and Eastern Polynesia. For example, haplogroup O3a2c, which has been previously linked to Taiwanese origins, is less abundant in East Polynesia. Similarly, Structure analyses based on autosomal markers indicate differences between West and East Polynesia. Yet, except for a European component in Nuku Hiva, no differences were detected in the Structure analyses between the Society and Marquesas Archipelagos. The Rst genetic distances also indicate a divide between Western and Eastern Polynesia, yet all pair wise comparisons between the islands of the Marquesas and Society archipelagos were not significantly different except for the Nuku Hiva-Hiva Oa combination. This lack of differentiation between the two East Polynesian archipelagos may be the result of the recent ancestral-descendent relationship and/or vigorous trade between the two sets of islands. A number of Melanesian, Polynesian and European Y-chromosome haplogroups exhibit different distributions between the Marquesan islands of Nuku Hiva and Hiva Oa. Although this phenomenon could have resulted from drift subsequent to colonization, it is possible that differential migration involving various ancestral populations and/or unique trading routes may have generated such distinctive patterns.
References
Wilmshurst JM, Hunt TL, Carl P, et al. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. PNAS. 2015;108:1815–20.
Mirabal S, Herrera KJ, Gayden T, et al. Increased Y-chromosome resolution of Polynesian Islands of Samoa and Tonga. Gene. 2012;492:339-48.
Gray RD, Drummond AJ, Greenhill SJ. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science. 2009;323:479–83.
Sagart, L. The expansion of Setaria farmers in East Asia: a linguistic and archaeological model. Past human migrations in East Asia: matching archaeology, linguistics and genetics. Routledge, NY, NY, USA. 2008;p. 133–57.
Stone R. Graves of the Pacific’s first seafarers revealed. Science. 2006;312:360.
Skoglund P, Cosimo Posth C, Sirak K. Genomic insights into the peopling of the Southwest Pacific. Nature. 2016;538:510–3.
Lipson M, Skoglund P, Spriggs M, et al. Population turnover in remote oceania shortly after initial settlement. Curr Biol. 2018;28(7):1157–65.
Posth C, Nägele K, Powell A. Language continuity despite population replacement in Remote Oceania. Nat Ecol Evol. 2018;2:731–40.
Bellwood P. The checkered prehistory of rice movement southwards as a domesticated cereal—from the Yangzi to the Equator. Rice. 2011;4:93–103.
Soares P, Trejaut JA, Loo JH, et al. Climate change and postglacial human dispersals in southeast Asia. Mol Biol Evol. 2008;25:1209–18.
Li H, Wen B, Chen S-J. Paternal genetic affinity between western Austronesians and Daic populations. BMC Evol Biol. 2008;8:146.
Tianlong J The neolithic of southeast China: cultural transformation and regional interaction on the coast. Amherst: Cambria Press; 2007.
Pawley A. The Austronesian dispersal: languages, technologies and people. In: Bellwood, PS, Renfrew C. Examining the farming/language dispersal hypothesis. UK: University of Cambridge Press; 2002.
Kayser M. The human genetic history of oceania: near and remote views of dispersal. Curr Biol. 2010;20:194–201.
Burley D, Weisler MI, Zhao J-X. High precision U/Th dating of first Polynesian settlement. PLoS ONE. 2012;7:e48769.
Hammarström H, Forkel R, Haspelmath M. Central Pacific. Glottolog 3.0. Jena, Germany: Max Planck Institute for the Science of Human History; 2017.
Vinton KP. The Lapita Peoples. London, UK: Wiley; 1997.
Hudjashov G, Endicott P, Post H, et al. Investigating the origins of eastern Polynesians using genome-wide data from the Leeward Society Isles. Sci Rep. 2018;8:1823.
Hunt TL, Lipo CP. Late colonization of Easter Island. Science. 2006;31:1603–6.
Horsburgh KA, McCoy MD. Dispersal, isolation, and interaction in the islands of Polynesia: a critical review of archaeological and genetic evidence. Diversity. 2017;9:37–58.
Green RC, Weisler MI. The Mangarevan sequence and the dating of geographic expansion into southeast Polynesia. Asian Perspectives. 2002;41:213–41.
Di Piazza A, Pearthree E. Voyaging and basalt exchange in the Phoenix and Line Archipelagoes: the viewpoint from three mystery islands. Archaeol Ocean. 2001;36:146–52.
Allen MS, William R, Dickinson WR, Huebert JM. The anomaly of Marquesan ceramics: a fifty year retrospective. J Pac Archaeol. 2012;3:90–104.
Sand C, Connaughton SP. Oceanic explorations: Lapita and western Pacific settlement, Australia: ANU E Press; 2007.
Stephanie L. Melanesia’: the history and politics of an idea. J Pac Hist. 2013;48:1–22.
Friedlaender JS, Friedlaender FR, Reed FA, et al. The genetic structure of pacific islanders. PLoS Genet. 2009;4:e19.
Kayser M, Lao O, Saar K, et al. Genome-wide analysis indicates more Asian than Melanesian ancestry of Polynesians. Am J Hum Genet. 2008;82:194–8.
Regueiro M, Mirabal S, Lacau H. Austronesian genetic signature in East African Madagascar and Polynesia. J Hum Genet. 2008;53:106–20.
Lansing JS, Murray P, Cox MP, et al. An ongoing Austronesian expansion in Island Southeast Asia. J Anthropol Archaeol. 2011;30:262–72.
Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, USA: Cold Spring Harbor Press; 1989.
Aznar JM, Celorrio D, Odriozola A, et al. I-DNASE21 system: development and SWGDAM validation of a new STR 21-plex reaction. Forensic Sci Int Genet. 2014;8:10–9.
Raymond M, Rousset F. genepop (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–9.
Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–106.
Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure using multilocus genotype data. Genetics. 2000;155:945–59.
Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20.
Nei M. Molecular evolutionary genetics. USA: Columbia University Press; 1987.
Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–62.
FMTA Busing, Commandeur JJF, Heiser WJ, Bandilla W, Faulbaum F, et al. PROXSCAL: a multidimensional scaling program for individual differences scaling with constraints. Adv Stat Softw. 1997;6:67–73.
Cardoso S, Zarrabeitia MT, Valverde L, et al. Variability of the entire mitochondrial DNA control region in a human isolate from the Pas Valley (northern Spain). J Forensic Sci. 2010;55:1196–201.
Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147.
Parson W, Gusmão L, Hares DR, et al. DNA Commission of the International Society for Forensic Genetics: revised and extended guidelines for mitochondrial DNA typing. Forensic Sci Int Genet. 2014;13:134–42.
van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–94.
Valverde L, Illescas MJ, Villaescusa P. New clues to the evolutionary history of the main European paternal lineage M269: dissection of the Y-SNP S116 in Atlantic Europe and Iberia. Eur J Hum Genet. 2016;24:437–41.
Hammer M, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. pp
Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7.
Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. USA: W.H. Freeman; 1969.
Kayser M, Brauer S, Cordaux R, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–44.
Wei LH, Shi Yan S, Teo YY, et al. Phylogeography of Y-chromosome haplogroup O3a2b2-N6 revealspatrilineal traces of Austronesian populations on the eastern coastal regions of Asia. PLoS ONE. 2017;12:e0175080.
Yan S. CC Wang, H Li, et al. An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4. Eur J Hum Genet. 2011;19:1013–5.
Salmond A. Aphrodite’s island. Berkeley USA: University of California Press; 2010.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Palencia-Madrid, L., Baeta, M., Villaescusa, P. et al. The Marquesans at the fringes of the Austronesian expansion. Eur J Hum Genet 27, 801–810 (2019). https://doi.org/10.1038/s41431-019-0336-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-019-0336-2