Abstract
Genetic interaction is a crucial issue in the understanding of functional pathways underlying complex diseases. However, detecting such interaction effects is challenging in terms of both methodology and statistical power. We address this issue by introducing a disease-concordant twin-case-only design, which applies to both monozygotic and dizygotic twins. To investigate the power, we conducted a computer simulation study by setting a series of parameter schemes with different minor allele frequencies and relative risks. Results from the simulation study reveals that the disease-concordant twin-case-only design largely reduces sample size required for sufficient power compared to the ordinary case-only design for detecting gene–gene interaction using unrelated individuals. Sample sizes for dizygotic and monozygotic twins were roughly 1/2 and 1/4 of sample sizes in the ordinary case-only design. Since dizygotic twins are genetically similar as siblings, the enriched power for dizygotic twins also applies to affected siblings, which could help to largely extend the application of the powerful twin-case-only design. In summary, our simulation reveals high value of disease-concordant twins and siblings in efficiently detecting gene-by-gene interactions.
Similar content being viewed by others
Introduction
Genome-wide association studies (GWAS) have helped identify genetic variations associated with various complex phenotypes [1,2,3]. However, single-nucleotide polymorphisms (SNPs) often only explain a limited fraction of the phenotype variance. On the other hand, epistasis or the genetic interactions could help with further improving the quantification of the genetic determinants of complex diseases and phenotypes [4,5,6]. Gene-by-gene interactions are often difficult to detect due to the computational challenge arising from the exponential growth of multilocus genotype combinations and large sample sizes required [7].
The case-only design has proven to be an efficient tool to detect gene–environment interaction [8, 9] and gene–sex interaction [10], and this powerful design has shown to be more efficient compared to the case–control design [11, 12]. In addition, the case-only design uses only cases and makes it more cost-effective in detecting interactions. The case-only design is also a valid approach to measuring the effects of gene–gene interactions [13] under the condition that the genes involved are in linkage equilibrium, i.e., alleles at the interacting loci are not inherited together.
Twins are special samples for genetic studies because of their genetic similarities and rearing-environmental sharing. The last century witnessed successful uses of twins in dissecting the genetic and environmental contributions to human diseases and complex traits. By comparing phenotype correlation patterns in monozygotic (MZ, identical) and dizygotic (DZ, fraternal) twin pairs, various genetic and environmental components can be assessed using the classical twin design for heritability estimation and correlation calculation [14]. With the advancement in biotechnology for genomic analysis, the use of twins is expanding from traditional genetic epidemiology to omics studies [15]. Through computer simulation, we have revealed the power advantage in using concordant twins in genetic association studies on human longevity and on complex diseases [16, 17]. In this study, we focus on analyzing gene–gene interactions by applying the powerful case-only design to affected twin pairs, the disease-concordant twin-case-only design, for more efficient detection of epistasis in complex diseases, assuming genetic interactions are enriched in disease-concordant twin pairs. The same idea can be extended to estimate gene-by-sex and gene-by-environment interactions.
Method
Experiment design
Instead of collecting patients from unrelated samples in an ordinary case-only design, we genotype and collect singletons from disease-concordant twin pairs in the twin-case-only design (Fig. 1). The design is flexible and could be applied to disease-concordant MZ, DZ twin as well as sibling pairs.
Simulation model
Assuming two SNPs (SNPA and SNPB) are under linkage equilibrium and for simplicity, each has a dominant effect. SNPA and SNPB, each has three genotypes: AA, Aa, aa and BB, Bb, bb, where a and b are minor alleles for SNPA and SNPB, respectively. Denote the minor allele frequencies for SNPA and SNPB as qA and qB. Then genotype frequencies for AA, Aa, and aa are (1 − qA)2, 2qA (1 − qA), qA2. Similarly genotype frequencies for BB, Bb, and bb are (1 − qB)2, 2qB (1 − qB), qB2. In total, there are nine multilocus genotypes for SNPA and SNPB. In the dominant mode of inheritance, genotypes Aa and aa have the same risk, therefore, the three genotypes of each SNP can be represented as two carrier types, and there are totally four multilocus carrier types (MLCTs). If we encode carrier as 1 and non-carrier as 0 for each SNP, and denote pij as MLCT frequency, where i, j are carrier types for SNPA and SNPB, we could compute these MLCT frequencies.
We define disease prevalence K as the proportion of individuals with the disease of interest in a population. Disease prevalence K was set to 0.05 in our simulation. Disease prevalence in a subpopulation of certain MLCT is denoted as dij, where i and j are carrier types for SNPA and SNPB, respectively. Then K is the weighted sum of disease prevalences for each MLCT [18].
When only interaction effect is considered, an individual carrying both SNPs has higher chance of developing disease. Then we could quantify the prevalence as d11 = rd10 = rd01 = rd00, where r is the relative risk. Combined with Eq. 2, we will have all disease prevalences shown in Eq. 3 for use in the simulation.
Simulating multilocus carrier type and phenotype
For unrelated individuals, we can simulate MLCTs and phenotypes with frequencies computed in Eqs. 1 and 3, respectively. Simulation of MLCTs in disease-concordant twin pairs is more complicated due to the relatedness between twin pairs. For MZ twin pairs who share same genotype, we simulate one MLCT and assigned it to the other twin of the same pair. For DZ twin pairs whose genotypes are possibly different, we start with the genotype simulations of their parents. Genotypes of SNPA and SNPB for each parent are simulated independently, and two alleles for each SNP have equal chance passing to their children to ensure that twins have 50% chance of inheriting an allele identical by descent (IBD) [16]. Phenotype of twins can be simulated independently using their genotypes and corresponding disease prevalences (Eq. 3), and we take one twin from each disease-concordant twin pair to form our study sample. To help accelerate the simulation process, we computed the conditional probabilities of different MLCTs. The computation of these probabilities can be found in Supplement Document 1.
Statistical testing and power estimation
We used Fisher’s exact test for both twin-case-only and ordinary case-only designs. Before we apply the test, a 2 × 2 contingency table with carrier types of two SNPs was summarized, and each cell represents number of individuals of corresponding MLCT. Then p-value of Fisher’s exact test was extracted for power estimation. The test was with 1 degree of freedom.
We simulated N = 2000 replicates for all scenarios, and corresponding powers were estimated as
where I [·] is an indicator function with 1 as true and 0 as false for logical expression of p <threshold. In our simulation, we are interested in testing one SNP that could be interacted with other SNPs in a GWAS, and thus we set p-value threshold as 5 × 10−8.
Although the description of simulation was on interacting SNPs, under the assumption that the interacting SNPs are in linkage equilibrium, our power estimates are also valid for detecting gene-by-sex and gene-by-environment interactions because the interacting factors can be coded as binary (0, 1) same as allele carrier status. Here the minor allele frequency can be replaced by the lower proportion of one sex or the proportion of exposures to an environmental condition.
Results
Empirical type I error rate
Empirical type I error rates were estimated prior to power simulation by setting relative risk r to 1. In the simulation, we set theoretical type I error rates to 0.05. We used different combinations of minor allele frequency of the two SNPs (0.05 and 0.30) in this simulation, and a sample size of 10,000 was used for each of a total 10,000 replications to ensure the accuracy of type I error rates. From the results shown in Table 1, we could see that all the empirical type I error rates are around 0.05, which suggests that the simulation and testing methods used were unbiased and our power simulation was valid.
Simulated power
The simulation was carried out with sample size increment of 10, and simulation stops when the power reaches 1 for each parameter set. Minor allele frequencies of both SNPs are the same as in empirical type I error rate simulation (Table 1), and relative risk r was set ranging from 1.2 to 2.5. The power estimates for comparing different designs were shown in Fig. 2. From the figure, we could see that power is stably increasing with sample size, and higher power is observed with higher frequency SNPs and higher relative risks. Most importantly, the highest power is achieved by the MZ twin-case-only design followed by the DZ twin-case-only design with ordinary case-only design showing the lowest power. For example, when qA = 0.05, qB = 0.3, and r = 1.25, power of MZ twin-case-only design reaches 80% when sample size is 1620. With same sample size, the power estimate for DZ twin-case-only design is 0.38 and for ordinary case-only design is 0.27. When sample size is 4160, power for DZ twin-case-only design is over 80%, and for ordinary case-only design, the power is only 0.58.
Power estimates for ordinary case-only design and twin-case-only design. Different line colors show different case-only designs. Each small plot is for a unique combination of different minor allele frequencies and relative risks. The horizontal red line marks power = 80%. qA minor allele frequency for SNPA, qB minor allele frequency for SNPB, r relative risk. (color figure online)
In Table 2, we also report minimal sample sizes required for power achieving 80%. Under the same setting of q and r and compared with the ordinary case-only design, the minimum sample size required can be roughly reduced by 1/2 sample size using the DZ twin-case-only design and by 1/4 using the MZ twin-case-only design. For example, when qA = 0.05, qB = 0.30, and r = 1.50, the estimated sample sizes are 1980, 1130, 430 for ordinary case-only, DZ and MZ twin-case-only designs, respectively. From Table 2, we also observe that the sample size estimated for power achieving 80% is very sensitive with regard to relative risk. For example, for qA = 0.05, qB = 0.05, when relative risk change from 1.20 to 1.25, estimated sample size changes from 27,410 to 18,250 for ordinary case-only design, which is over 1/3 change. With disease-concordant twins, those numbers are from 16,950 to 10,820 by the DZ twin-case-only design and from 6330 to 4120 by the MZ twin-case-only design.
Illustration with empirical data
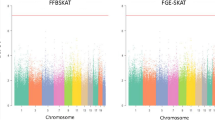
In addition to the simulated results, we further illustrate the high efficiency of the disease-concordant twin-case-only design in detecting gene-by-sex interaction using an empirical data set from a GWAS project based on 363,536 genotyped SNPs and a total of 1,348,667 SNPs after imputation and quality control [19]. The project collected 434 patients with allergic rhinitis consisting of 243 siblings and 191 unrelated individuals, both male and female. Although with small sample sizes, the presence of both sibling pairs who are genetically correlated as DZ twins and unrelated individuals in the sample offers us an opportunity to perform sibling-case-only and ordinary/unrelated case-only analyses and compare their performances in detecting gene–sex interaction. To do that, we used two subsets from the sample, 138 siblings and 138 unrelated individuals, and we matched both sample size and gender proportion. For each of the two datasets, we conducted a GWAS using logistic regression to test genotype by sex dependence.
The sibling-case-only design identified 5 SNPs with p-value <1 × 10−5 and 82 SNPs with p-value below 1 × 10−4. While in the ordinary case-only design, no SNP is found with p-value below 1 × 10−5 and only 13 SNPs had p-value below 1 × 10−4. To ensure that no inflation in the statistical significance is involved, we calculated genomic inflation factor (GIF) [20], with GIF = 1.017 for the disease concordant sibling-case-only design and GIF = 1.011 for the ordinary case-only design. Since the two GIFs are nearly equal and are both very close to 1, there is no genomic inflation in the two GWASs. Although no significant hit was found for both designs, the fact that more SNPs were found with p-value below relatively low or suggestive thresholds (1 × 10−5 and 1 × 10−4) by the sibling-case-only design in combination with its GIF of about one suggests that it is more sensitive and efficient compared to the ordinary case-only design.
Discussion
Despite the recent success of GWAS in identifying loci associated with complex diseases, a large proportion of the genetic components remain missing perhaps partly due to the ignorance of genetic interactions in the current GWAS. Detecting gene–gene, gene–sex, and gene–environment interactions might help with improving the quantification of the genetic determinants of complex diseases and phenotypes. As testing for the interaction effects in genomic association analysis is a challenging frontier, the simulation results on our proposed disease-concordant twin-case-only design for detecting interaction effects are encouraging. With a roughly 1/2 sample size reduction by the DZ and 1/4 with MZ twin-case-only designs as compared with the ordinary case-only design, our proposed approach is a powerful tool for detecting interaction effects in genome-wide association analysis.
The fact that disease concordance occurred in genetically related twin pairs or siblings highly increases the probability of sharing disease genes including interacting genes, with the higher the degree of genetic relatedness the higher the probability of sharing. We want to mention here that the disease-concordant twins/siblings design for association analysis also benefits from the power linkage analysis because the sharing of interacting genes in MZ twins is actually IBD from a common ancestor without any intervening recombination. Even in disease-concordant DZ twins or siblings, the sharing can be IBD or identical-by-state. This means that the disease-concordant twin design tests not only genetic association, but also, to some extent, genetic linkage as in the affected sib-pair analysis [21]. The highly enriched power by applying the case-only design to disease-concordant twins could offer a new opportunity for exploring the genetic interaction network that involves genes of low frequency and/or small effect size, which would be hard to detect using conventional methodology [22].
As just aforementioned, the enriched power of the proposed design is due to the increased likelihood of interaction effects in genetically related individuals who are affected, if genetic interaction is indeed involved in disease development. This also imposes restriction on sample collection, which could limit practical application of the design. Fortunately, the same power advantage in using disease-concordant DZ twins also applies to affected sibling pairs due to the same degree of genetic sharing. Extending to the sampling scope to include affected sibling pairs largely increases feasibility for studies using the proposed design. On the other hand, the frequent use of twins in genomic studies also offers new opportunities for conducting large-scale, consortium-based analysis of epistasis and gene–sex as well as gene–environment interactions in GWAS. Finally, considering the high experimental expenses in genomic analysis, promoting the disease-concordant twin-case-only design is a cost-effective way for uncovering the interactive network in disease development.
Finally, the power estimates presented in this paper are for carriers of SNP minor alleles (non-carriers coded 0, carriers coded 1) with power simulated using a p-value cutoff for GWAS. Because of that, the power estimates also apply to tests on gene-by-sex (as illustrated in the example application) and gene-by-environment interactions by coding sex and exposure as 0 or 1, with the degree of multiple testing similar to a typical GWAS. This flexibility expands the application of the twin-case-only design to cover different types of interactions involving the genome to further harness the power of twins in genomic studies.
References
Visscher PM, Wray NR, Zhang Q, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–83.
Price AL, Spencer CCA, Donnelly P. Progress and promise in understanding the genetic basis of common diseases. Proc R Soc B. 2015;282:20151684.
Onay VÜ, Briollais L, Knight JA, et al. SNP-SNP interactions in breast cancer susceptibility. BMC Cancer. 2006;6:1–16.
Dinu I, Mahasirimongkol S, Liu Q, et al. SNP-SNP interactions discovered by logic regression explain Crohn’s disease genetics. PLoS ONE. 2012;7:e43035.
Jamshidi M, Fagerholm R, Khan S, et al. SNP-SNP interaction analysis of NF-κB signaling pathway on breast cancer survival. Oncotarget. 2015;6:37979–94.
Gilbert-Diamond D, Moore JH. Analysis of gene-gene interactions. Curr Protoc Hum Genet. 2011;70:1.14.1–12.
Gatto NM, Campbell UB, Rundle AG, Ahsan H. Further development of the case-only design for assessing gene-environment interaction: evaluation of and adjustment for bias. Int J Epidemiol. 2004;33:1014–24.
Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–62.
Tsuchiya M, Iwasaki M, Otani T, et al. Breast cancer in first-degree relatives and risk of lung cancer: assessment of the existence of gene sex interactions. Jpn J Clin Oncol. 2007;37:419–23.
Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404.
Hassanzadeh J, Moradzadeh R, Fard AR, Tahmasebi S, Golmohammadi PA. Comparison of case-control and case-only designs to investigate gene-environment interactions using breast cancer data. Iran J Med Sci. 2012;37:112–8.
Yang Q, Khoury MJ, Sun F, Flanders WD. Case-only design to measure gene-gene interaction. Epidemiology. 1999;10:167–70.
Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3:872–82.
Tan Q, Kyvik KO, Kruse TA, Christensen K. Dissecting complex phenotypes using the genomics of twins. Funct Integr Genomics. 2010;10:321–7.
Tan Q, Zhao JH, Kruse T, Christensen K. Power estimation for gene-longevity association analysis using concordant twins. Genet Res Int. 2014;2014:8.
Tan Q, Li W, Vandin F. Disease-concordant twins empower genetic association studies. Ann Hum Genet. 2017;81:20–6.
Tan Q, Zhao JH, Zhang D, Kruse TA, Christensen K. Power for genetic association study of human longevity using the case-control design. Am J Epidemiol. 2008;168:890–6.
Mohammadnejad A, Brasch-Andersen C, Li W, Haagerup A, Baumbach J, Tan Q. A case-only genome-wide association study on gene-sex interaction in allergic rhinitis. Ann Allergy Asthma Immunol. 2018;121:366–7.
Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66:1933–44.
Wu C, Amos CI. Statistical properties of affected sib-pair linkage tests. Hum Hered. 2003;55:153–62.
Murk W, Dewan AT. Exhaustive genome-wide search for SNP-SNP interactions across 10 human diseases. G3. 2016;6:2043–50.
Acknowledgements
This study was jointly supported by the Lundbeck Foundation (grant number R170-2014-1353) and the DFF research project 1 from the Danish Council for Independent Research, Medical Sciences (DFF-FSS): DFF-6110-00114 and DFF-6110-00016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, W., Baumbach, J., Mohammadnejad, A. et al. Enriched power of disease-concordant twin-case-only design in detecting interactions in genome-wide association studies. Eur J Hum Genet 27, 631–636 (2019). https://doi.org/10.1038/s41431-018-0320-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0320-2
This article is cited by
-
Genome-wide DNA methylation analysis of pulmonary function in middle and old-aged Chinese monozygotic twins
Respiratory Research (2021)