Abstract
ST3GAL3 encodes the Golgi enzyme beta-galactoside-alpha-2,3-sialyltransferase-III that in humans forms, among others, the sialyl Lewis a (sLea) epitope on proteins. Functionally deleterious variants in this gene were previously identified in patients with either non-syndromic or syndromic intellectual disability such as West syndrome, an age-dependent epileptic encephalopathic syndrome associated with developmental arrest or regression. The aim of this study was to further elucidate the molecular and cellular mechanisms causing West syndrome by lack of ST3GAL3 function. For this purpose we generated induced pluripotent stem cell (iPSC) lines from fibroblasts obtained from a patient with West syndrome, carrying a variant in exon 12 (c.958G>C, p.(Ala320Pro)) of ST3GAL3, and a healthy sibling, using lentiviral reprogramming. iPSCs and cortical neurons derived thereof were analysed by lectin blots, mRNA sequencing, adherence assays, and FACS. While no significant difference was observed at stem cell or fibroblast level between patient and control cells, patient-derived cortical neurons displayed an altered lectin blot staining pattern, enhanced adherence to a poly-l-ornithine/laminin-coated surface and decreased levels of neurons expressing T-box transcription factor brain 1. Our results suggest that changes in the sialylation pattern on the surface of specific neuronal cell types affect adhesive interactions during development, which in turn may cause subtle changes in tissue composition that could result in the occurrence of epilepsy and might impair neural development to an extent that is detrimental to the development and maintenance of normal cognitive functions.
Similar content being viewed by others

Introduction
Intellectual disability (ID) is a cognitive impairment disorder characterized by an intelligence quotient (IQ) below 70 and is one of the most common forms of cognitive handicaps resulting from genetic aberrations [1]. Hereditary forms of ID are genetically heterogeneous, but a considerable number involve genes participating in the build-up and maintenance of the glycocalyx [2]. The glycocalyx is composed of carbohydrates that are part of the cell surface-exposed lipids and proteins (including proteoglycans) or part of the extracellular matrix (ECM). The term glycan is used to refer to these complex carbohydrates and to differentiate them from metabolically used sugars. At the interface to the outside world, glycans form the major cellular communication structures and are involved in the regulation of cell–cell interactions steering inter alia cell adhesion and migration [3]. In model organisms the ablation of genes that encode central enzymes in glycosylation pathways (henceforth referred to as “glycogenes”) frequently cause embryonic lethality [4]. In contrast, hypomorphic variants in glycogenes remained unrecognized for a long time because these rare gene defects cause multi-systemic disorders now summarized as congenital disorders of glycosylation (CDGs) [2].
We recently demonstrated that variants in ST3GAL3 (NM_006279.2), which encodes the Golgi enzyme beta-galactoside-alpha-2,3-sialyltransferase-III, lead to varying clinical manifestations. Two independent variants, c.38C>A (p.(Ala13Asp) LOVD [5] variant ID: 0000357720 (http://www.lovd.nl/ST3GAL3)) and c.1108G>T (p.(Asp370Tyr) LOVD [5] variant ID: 0000357721 (http://www.lovd.nl/ST3GAL3)), were associated with mild forms of non-syndromic autosomal recessive intellectual disability (NSARID) [6]. Ectopic expression of full-length-c-MYC fusion proteins with these variants in LMTK-- cells showed severe functional impairment and subcellular mislocalization of both mutant gene products [6]. A third variant, c.958G>C (p.(Ala320Pro) LOVD [5] variant ID: 0000357719 (http://www.lovd.nl/ST3GAL3)) in exon 12 of the gene product, identified in a Palestinian family caused West syndrome (WS), an age-dependent epileptic encephalopathic syndrome associated with developmental arrest or, as in the case of our patients [7], regression [8]. The exon numbering applied here (according to RefSeq NM_006279.4; Gencode Transcript: ENST00000361392.8; Gencode Gene: ENSG00000126091.19; Protein: Q11203) starts with the exon containing the transcription start site at chr1 pos. 43707497 bp (GRCh38/hg38) as exon number 1.
Human ST3GAL3 is highly expressed in the developing brain [9] and was demonstrated to form predominantly sialyl Lewis a (sLea) epitopes on proteins [10]. The latter are known to be crucially involved in the regulation of cell–cell interactions [11]. Interestingly, a second member of the ST3GAL subgroup, ST3GAL5, which is involved in ganglioside synthesis [12], has been reported to play a role in the aetiology of ID disorders involving epilepsy [13, 14].
A major challenge in understanding the molecular basis of human brain function is to link genotype and phenotype. This task is even more challenging as primarily affected tissue is inaccessible to trace back alterations resulting from gene defects. However, being in the fortunate situation of having access to specimens obtained from the WS patient [7] and her healthy sibling, we were able to take advantage of the option to generate induced pluripotent stem cells (iPSCs) from patient and control fibroblasts to perform comparative analysis of neural differentiation starting from pluripotent stem cells. After 50 days of succsesful neuronal differentiation, patient-derived cultures showed a reduced number of T-box transcription factor brain 1 (TBR1)-positive cortical projection neurons and altered adherence properties. Thus, our results indicate that ST3GAL3 function is important for the normal development and functioning of the brain.
Materials and methods
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
An extended version of this section with more specific experimental details is provided as supporting material.
Mutagenesis and overexpression of ST3GAL3-c-MYC fusion proteins in LMTK− cells
To introduce variants of interest into ST3GAL3, we used the QuikChange site-directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA) according to the manufacturers’ instructions. Mutant and wild-type ST3GAL3 were expressed as fusion proteins with C-terminally added c-MYC in LMTK− cells.
Patients and cell lines
Detailed information about primary human cell lines and patient material is given in Supplementary Table 2. For transcriptome analysis of normal human brain tissue, commercially available RNA preparations were used (Supplementary Table 3).
Fibroblast culture and transfection
For reprogramming into iPSCs, fibroblasts from a female patient homozygous for the ST3GAL3 variant c.958G>C and her female healthy sibling were obtained in accordance with the relevant ethical guidelines and approved by the appropriate institutional review board. To culture iPSCs, mouse embryonic fibroblasts (MEFs) were used as feeders (MEF (C57BL/6) (MEF-BL/6-1), SCRC-1008™, ATCC, USA). Studies on the subcellular localization of wild-type and mutant ST3GAL3 were carried out in LMTK-- cells. For fibroblast culture, culture dishes were coated with 0.1% (w/v) gelatine. Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal calf serum, 100 units/ml penicillin, and 100 µg/ml streptomycin was used as culture medium. Cells were kept at 37 °C in the presence of 5% CO2.
Prior to use, murine fibroblasts were treated with 10 µg/ml mitomycin. LMTK-- cell transfections and expression of ST3GAL3 fusion proteins were performed as previously described [7].
Reprogramming of fibroblasts
The reprogramming workflow is shown in Supplementary Figure 1A. A self-inactivating lentiviral vector carrying the reprogramming factors OCT4, KLF4, SOX2, and c-MYC was used and applied as described recently [15]. Fibroblasts were seeded at a density of 30,000 cells/cm [2] and transduced with viral particles at a multiplicity of infection of 0.25.
Human iPS cell culture and differentiation
For culture of iPSCs we applied two methods: (i) culture on MEFs as feeder cells and (ii) culture on MatrigelTM (BD Bioscience, Franklin Lakes, NJ, USA).
For neuronal differentiation, iPSCs were cultured on Matrigel in MEF-conditioned media (CM). At confluency, MEF-CM was replaced by N2/B27 medium supplemented with 1 µM dorsomorphin (RnD Systems Inc., Minneapolis, MN, USA), 10 µM SB431542 (Millipore, Billerica, MA, USA), and retinoic acid (50 µg/µl).
For cortical differentiation (Supplementary Figure 2A) we adapted the protocol developed by ref. [16].
Quantitative real-time PCR
TriZol (Life Technologies, Carlsbad, CA, USA) was used to isolate total RNA. For complementary DNA (cDNA) analysis, 250 ng total RNA was reverse transcribed using random hexamer primers. Real-time PCR (RT-PCR) was performed in 20 µl reaction volume with 12.5 ng cDNA each. Primers were intron spanning. Amplification and analysis was performed with DNA SYBR Green I master on a Light Cycler 480 (Roche, Basel, Switzerland).
Fusion primer and nested integrated PCR (FPNI PCR)
The virus cassette integration site was located using fusion primers and nested integrated PCR according to ref. [17]. The primer for the first amplification was designed to bind within the truncated long terminal repeats of the virus cassette.
Array comparative genomic hybridization (Array-CGH)
Cells were arrested at G1/G0 by adding 100 nM Torin-1 (RnD Systems Inc., Minneapolis, MN, USA) to the culture for 5 h and subsequently harvested. For DNA isolation the NucleoSpin Tissue Kit (Marchery & Nagel, Düren, Germany) was used. Array-CGH was performed as described previously [15].
Immunofluorescence staining
Cells were cultured on sterile, coated cover glasses, fixed with para-formaldehyde, permeabilized using Triton X-100, and blocked with normal goat serum. Incubations with primary antibodies were carried out overnight at 4 °C and with secondary antibodies for 1 h at room temperature. Antibodies and dilutions are listed in Supplementary Table 3.
RNA-sequencing
Gene expression analysis by RNA-sequencing was performed as described previously using a SOLiD 5500XL instrument (Fisher Scientific, Carlsbad, CA, USA) [18]. Read mapping (GRCh37/hg19) was performed using the Lifescope software (Life Technologies, Carlsbad, CA, USA). Bioinformatic analyses were performed using the R package DESeq [19].
Lectin/western blots
Cells were cultured on MatrigelTM or poly-l-ornithine/laminin-coated culture dishes and harvested with 0.5 µM EDTA solution. Cell lysis, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), western blot, and lectin staining was performed as described previously [15]. Lectins and dilutions are listed in Supplementary Table 4.
Cell adherence assay
Cells were washed twice in 1× phosphate-buffered saline, incubated in 0.5 µM EDTA solution for 7 min at 37 °C and harvested in DMEM (Life Technologies, Carlsbad, CA, USA) basal media. The suspension was spun down for 5 min at 250 × g and the supernatant discarded. Cell pellets were resuspended in medium and cells counted using a Neubauer chamber. A defined number of cells was then seeded onto gelatine or poly-l-ornithine/laminin-coated 12-well culture dishes and re-incubated at 37 °C (5% CO2). For the fibroblast adherence assay, supernatants were removed after 5, 10, 20, and 40 min. To determine the adherence velocity of cortically differentiated cells (30 days), the supernatants were removed after 2, 5, 10, and 30 min. Cell numbers in the supernatants were counted with the help of a Neubauer chamber and subtracted from the number of seeded cells to give a measure of adherence velocity.
Bioanalyzer “Cell assay”
Immunocytochemistry and counting of positively stained cells was performed following the manufacturers’ instructions for the Agilent “Cell Kit” (Agilent, Santa Clara, CA, USA). To distinguish live cells from dead cells, all cells were stained with SYTO 16 (Life Technologies, Carlsbad, CA, USA), fixed with para-formaldehyde solution, and permeabilized with Triton X-100 solution. Primary antibodies and allophycocyanin-labelled secondary antibodies were incubated in the dark. For analysis, cells were resuspended in ABI cell buffer at a density of 2 × 106 cells/ml.
Results
To provide an overview over the features that are conserved in the vertebrate ST-family, a schematic representation of ST3GAL3 is given in Fig. 1a. Amino acid positions that were previously found to be mutated in NSARID [6] and WS patients [7] (red box) are indicated and henceforth used to denominate the respective cDNAs and proteins.
ST3GAL3 structure and localization. a Like other STs, ST3GAL3 is a Golgi-localized type II membrane protein, characterized by a short NH2-terminal cytosolic domain, an amphipathic transmembrane domain (TMD) followed by a stem region, and a COOH-terminal catalytic domain. The latter contains the highly conserved sialyl motifs large (L), short (S), motif III (III), and very short (VS). Individual (black triangles) as well as conserved (red triangles) N-glycosylation sites are shown, Amino acid positions found to be mutated in the NSARID (black letters [6]) and WS patients (red frame [7]) are indicated. b Immunofluorescence images of LMTK-- cells ectopically expressing ST3GAL3 variants as c-MYC fusion proteins. C-MYC (“anti-MYC”, green) and Golgi marker giantin (“anti Giantin”, red) staining with specific primary antibodies (aGia, Covance, PRB-114C, 1:1000) and corresponding secondary antibodies (aMs-Alexa Fluor 488, Life Technologies, A-11029, 1:200; aRb-Alexa Fluor 555, Life Technologies, A-21429, 1:600) is shown for wild-type (wt) ST3GAL3 and the three known disease-causing variants. The red frame indicates the variant Ala320Pro, the focus of this report
Because the ectopic expression of full-length-c-MYC fusion proteins in LMTK-- cells led to a subcellular mislocalization of ST3GAL3Ala13Asp and ST3GAL3Asp370Tyr as shown in previous studies [6], we first examined the localization of mutant ST3GAL3Ala320Pro [7], which harbours a variant in the functionally important sialyl-motif S. The expression vector pcDNA3.1/MYC-hisA-ST3GAL3wtwas used as template to generate MYC-hisA-ST3GAL3Ala320Pro by site-directed mutagenesis. With LMTK-- cells as hosts both variants were expressed and visualized by immunostaining. While the ST3GAL3wt-c-MYC signal co-localized with the Golgi marker giantin, transfection of cells with MYC-hisA-ST3GAL3Ala320Pro generated a diffuse endoplasmic reticulum (ER)-like staining (Fig. 1b), indicating ER retention of MYC-hisA-ST3GAL3Ala320Pro, which is in perfect agreement with our previous observations for the variants ST3GAL3Ala13Asp and ST3GAL3Asp370Tyr [6].
Generation of a ΔST3GAL3 iPSC model
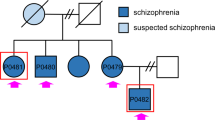
Fibroblasts from the patient homozygous for the ST3GAL3Ala320Pro variant (4695; ΔST3GAL3-HF) [7] and a healthy sibling (4696; “AS”) homozygous for the wild-type allele, and thus providing a near isogenic control, were reprogrammed by transduction with a multicistronic lentiviral vector harbouring four transcription factors (OCT4, SOX2, KLF4, and c-MYC) and the reporter dye dTomato [7, 20]. Successful transduction was monitored by red fluorescence of the reporter dye. Within 2 weeks after transduction, the majority of cells had begun changing from typical spindle-like fibroblast morphology to a stem cell-like appearance characterized by a round cell shape and a high nucleus to cytoplasm ratio. At day 20 post transduction, these cells had grown to stem cell-like colonies with sharp borders. Silencing of exogenous transcription factor expression was displayed by the absence of the reporter dye signal (Supplementary Figure 1). Reprogramming of both patient and control fibroblasts yielded clones that were isolated, expanded, and assessed for genomic integrity and pluripotency. To investigate genomic integrity after reprogramming, nested integrated PCR with fusion primers was performed to detect the number and exact genomic position of virus cassette integration [17]. The ΔST3GAL3 clone 1 displayed two integration sites, one at position 6.956.876 (GRCh37/hg19) on the short arm of chromosome X and one at position 25.663.792 on the long arm of chromosome 17. AS clone 23 (ASC23) showed one integration site at position 158.904.392 (GRCh37/hg19) on the long arm of chromosome 4. Neither of these integration events was in or near to an open reading frame. Overall genomic integrity was analysed with ΔST3GAL3 clone 1, ASC23 and their corresponding fibroblasts by array-CGH. Neither of the clones showed copy number alterations when compared to the respective fibroblast cell line of origin (data not shown). To validate pluripotency of the generated iPSC lines, expression of six pluripotency-associated genes (LIN28A, NANOG, OCT4, PODXL, REX1, and SOX2) [21] were validated by quantitative real-time PCR (RT-qPCR). Comparison of RT-qPCR data with total RNA-sequencing data obtained for the two cell lines and human embryonic stem cells confirmed similar expression levels in all six genes, with LIN28a, PODXL, and DNMT3B displaying the highest expression levels (Supplementary Figure 1C). In addition, expression of OCT4 and the pluripotency-associated glycosphingolipid SSEA4 was verified by immunofluorescence staining (Supplementary Figure 1D). Moreover, comparison of RNA-sequencing data of patient and healthy sibling-derived iPSCs showed only few genes with significantly different expression levels (Supplementary Figure 4A). These results were supported by principal component analysis of both data sets with a third unrelated iPSC line and an embryonic stem cell line, showing that patient and sibling cell lines were much more similar to each other than were the cells from unrelated individuals (Supplementary Figure 4B). Altogether, these results indicate that both iPSC lines, derived either from the WS patient or its healthy sibling, show as little differences in their RNA expression profile as expected for a near isogenic control.
Cortical differentiation
In addition to severe ID, individuals homozygous for ST3GAL3Ala320Pro had epileptic seizures [7]. As seizures primarily originate in cortical brain regions [22] and the ST3GAL3Ala320Pro homozygous patient did not respond to antiepileptic treatment [7], we aimed to identify the brain region with highest ST3GAL3 expression. RNA-sequencing of different adult human brain areas was carried out and showed highest expression levels for ST3GAL3 in cortical samples (Supplementary Table 2). Therefore, we adapted a differentiation protocol (modified from Shi et al. [23]) that allowed the development of cortical neurons and neuronal networks, with the exception of interneurons.
After neural induction, both lines displayed all appropriate developmental stages including neuroectoderm (days 9–12), neural progenitor cells arranged in typical rosette-like structures (from day 15) [24], and first neurons evolving at the outside of rosette structures (from day 20; Supplementary Figure 2A,B). Cortical identity was validated by RT-qPCR analysis of cortical markers (FOXG1, EMX1, SOX1, OTX1, and PAX6) on cortically differentiated cells [23] after 20 days. The messenger RNA (mRNA) expression levels of these markers were compared to those in undifferentiated iPSCs. All samples showed increased expression of cortical markers and decreased expression of the pluripotency marker OCT4 (Supplementary Figure 2C). In addition, the cortical marker OTX1 and the neural progenitor marker PAX6 were analysed by immunofluorescence staining, confirming the qPCR results (Supplementary Figure 2D). Taken together, these findings show that iPSCs derived from patient and healthy sibling could be differentiated into cortical cells with similar efficiency.
Biochemical characterization of mutant iPSCs and iPSC-derived cortical cells
To investigate the effect of ST3GAL3 deficiency on protein glycosylation we applied lectin staining of whole cellular proteins from patient- and healthy sibling-derived iPSCs as well as cortical cells obtained from these iPSCs after 50 and 90 days of differentiation, respectively, upon separation by SDS-PAGE. Staining with Maackia amurensis agglutinin (MAA), a lectin specific for sialic acids in α-2,3-linkage, did not reveal any differences between patient (ΔST3GAL3) or control (ASC23) iPSCs (Fig. 2a, upper left panel). However after 50 and 90 days of cortical differentiation, a protein band at about 80 kDa was detected with slightly increased intensity in control compared to patient samples by MAA (Fig. 2a, upper right panel). On the other hand, applying lectin staining with Datura stramonium agglutinin (DSA), detecting β-1,3-linked galactose (the sugar below sialic acids), revealed that at the same height of about 80 kDa a protein band is detected with increased intensity in d50 and d90 differentiated samples derived from patient iPSCs compared to control cells (Fig. 2a, lower left panel). Staining with Sambucus nigra agglutinin (SNA), a lectin detecting sialic acids in α-2,6-linkage, did not reveal any differences between patient and control samples upon cortical differentiation (Fig. 2a, lower right panel). Taken together, our findings imply that a protein of about 80 kDa is sialylated in d50 and d90 differentiated samples in α-2,3-linkage by ST3GAL3.
Lectin blots and cell adherence assays. a Lectin blots. The 5 µg protein of whole cell lysates of either undifferentiated iPSCs (d0) or upon 50 (d50) or 90 days (d90) of cortical differentiation from cells of the healthy sibling (ASC23) or the patient (ΔST3GAL3) were separated by SDS-PAGE, blotted onto nitrocellulose, and stained with lectins against terminal α-2,3-linked sialic acid (MAA), β-1,3-linked galactose (DSA), or α-2,6-linked sialic acid (SNA). Lectin binding is shown in grey scale. Blots were re-probed with a specific antibody against the housekeeping protein glycerinaldehyde 3-phosphate dehydrogenase (GAPDH) and antibody binding is shown in green. Arrows indictate a protein band at about 80 kDa. Solid and dashed lines highlight binding as well as less or no binding of lectins, respectively. PR PageRuler Prestained Protein Ladder (Thermo Fisher), pos positive control (1 µg fetuin), neg negative control (1 µg asialofetuin). b Adherence of cells obtained 30 days upon neural induction of iPSCs and c adherence of fibroblasts were analysed applying an adherence assay. The mean fractions of adherent cells over time are displayed. Means ± s.d. were calculated from three independent experiments, each performed in duplicate. Error bars represent the standard deviation
Cortical cells derived from ΔST3GAL3 clone 1 show elevated adherence velocity
Comparison of cortically differentiating cells from patient and healthy sibling showed no morphological differences. Neural progenitor cells formed rosette-like structures and evolving neurons had a typical triangular cell body with long dendrites (Supplementary Figures 2B and 3) as described previously [25]. In long-term cultures between day 50 and day 90 of differentiation, neural rosettes began to form very large and complex cortical structures resembling neural networks (Supplementary Figure 3). However, patient cells showed remarkably altered adherence during passaging. We further investigated this feature by cell adherence assays. Strikingly, quantification of adherence velocity revealed a significant increase for cortical neurons derived from patient iPSCs. Within 2 min, about 40% of ΔST3GAL3 iPSC-derived cortical neurons adhered, whereas in the same time only 16% of healthy control cells bound to the surface of the culture dish (Fig. 2b). In contrast to iPSC-derived neurons, the patient-derived fibroblasts containing the ST3GAL3 variant exhibited no altered adherence velocity (Fig. 2c).
Diminished TBR1+ neuron population in patient samples
Increased adherence velocity caused by ST3GAL3 deficiency may indicate a particular vulnerability for cell types that in vivo depend on a high degree of plasticity (motility). This could mean that such cell types are underrepresented in the (developing) brain of individuals with deleterious ST3GAL3 variants. Therefore, we next investigated if differences in the neuronal subtype compositions occur during in vitro differentiation.
The human cortex consists of six layers (laminae) (Fig. 3a). In vitro as well as in vivo, genesis of projection neurons follows a stereotypical inside-out manner [26]. After 35 days of in vitro differentiation, two progenitor populations of these projection neurons, PAX6-expressing radial glia cells and intermediate neuronal progenitor cells expressing TBR2 (T-box brain 2), can be expected. As shown in Fig. 3e, both populations were present at day 35. PAX6+ radial glia cells (shown in green), grouped into typical rosette-like structures, and TBR2+ intermediate neuronal progenitor cells (shown in red) were located at the periphery of these structures. Both progenitor populations contribute to the proliferation of the first emerging projection neurons by asymmetric cell division (Fig. 3d).
Projection neurons in the developing cortex. a Schematic illustration of projection neurons in the different cortical layers (modified after Shi et al. [23]). b Immunofluorescence analysis of patient (ΔST3GAL3) and healthy sibling (ASC23) cells after 50 days of cortical differentiation. An antibody against Tbr1 as marker for cortical projection neurons of lamina VI (Abcam, ab31940, 1:300; anti-rabbit IgG-Alexa Fluor 555, Life Technologies, 1:500; Secondary antibody: aRb-Alexa Fluor 555, Life Technologies, A-21429, 1:600) was used. Scale bars: 50 µM. c Quantity of projection neurons, measured after 50 days of cortical differentiation of patient (ΔST3GAL3) and healthy sibling (ASC23) cells. Immunofluorescence staining was done against Tbr1 (Abcam, ab31940, 1:100) or Satb2 (Abcam, ab51502, 1:50) using APC labelled secondary antibodies (Life Technologies, A-10931, 1:350, A-865, 1:100). All cells were stained with SYTO 16 (Life Technologies, S7578, 1 nM). Relative amounts of TBR1- or SATB2-positive cells in patient and healthy sibling are shown. Error bars represent the standard deviation (n = 9). d Schematic depiction of cortical stem cell differentiation. Radial glia cells (PAX6 positive, green) divide asymmetrically into intermediate progenitor cells (TBR2 positive, orange) or directly into projection neurons (TBR1 positive, red). e Immunofluorescence images of patient (ΔST3GAL3) and healthy sibling (ASC23) cells after 35 days of cortical differentiation, using antibodies against PAX6 (aPax6, SCB, SC-81649, 1:300, green) and TBR2 (aTbr2, Abcam, ab23345, 1:100, red) As secondary antibodies aMs-Alexa Fluor 488 (Life Technologies, A-11029, 1:200) and aRb-Alexa Fluor 555 (Life Technologies, A-21429, 1:600) were used respectively. Scale bars: 50 µM
TBR1 is a marker of early-born deep-layer (cortico-thalamic projection) neurons [27]. After 50 days of differentiation, TBR1-positive cells could be repeatedly detected in iPSC-derived cultures from the affected individual and from the control (Fig. 3b). However, when quantified, the number of TBR1+ neurons was drastically diminished in patient-derived cells (ΔST3GAL3, red, 23.88%) compared to healthy sibling-derived cells (ASC23, green, 54.51%). In order to test for an overall effect on neurogenesis, we also analysed the appearance of cells positive for SATB2, a marker for later-born neurons of the upper layers [28]. Because this analysis revealed no significant difference in numbers (5.65 and 4.54% of SATB2+ cells in the ASC23- and ΔST3GAL3-derived cultures, respectively, see Fig. 3c), we assume that in homozygous carriers of the deleterious ST3GAL3 variant the genesis of early-born TBR1+ cortical (cortico-thalamic projection) neurons is specifically affected.
Discussion
In this study, we provide evidence that a ST3GAL3 variant known to cause West syndrome in homozygous carriers [7] leads to subcellular mislocalization of the corresponding gene product, which is in keeping with our previous observations for two other ID-causing ST3GAL3 variants [6], suggesting that disease-causing ST3GAL3 variants result in an absence of this protein from the Golgi apparatus.
When comparing these variants with respect to their direct molecular effects, the Ala13Asp variant is partially mislocalized with slightly reduced enzymatic activity, and the Asp370Tyr variant is completely mislocalized but with residual enzymatic activity. Both cause ID without apparent epilepsia [6]. The Ala320Pro variant, which is the subject of this study, is completely mislocalized with no residual enzymatic activity and leads to the more severe phenotype of West syndrome [7]. Still, variations resulting in a proline incorporation can significantly affect the secondary and tertiary structure of proteins [29] as proline differs from all other naturally occurring amino acid residues in two important aspects: The phi dihedral angle of Pro is constrained to values close to −65 degrees (resulting in an extremely different angle in the primary and thus subsequent structures) and this amino acid also lacks an amide hydrogen. Consequently, mutations which result in an introduction of proline can significantly affect protein stability. It is therefore not surprising that the proline variation in the enzymatic active site of ST3GAL3 results in a total loss of enzymatic function and thus leads to more severe consequences for the affected individuals. Moreover, the expression of the other members of the ST3GAL family (RNA-sequencing results, data not shown) was in all cases unaltered between patient and control at both 50 and 90 days of neuronal differentiation of patient-derived iPS cells, so that any quantity-based rescue effect by closely related homologues can be ruled out.
iPSC-derived neuronal cell types are a promising model for the investigation of functional aspects of variants involved in the aetiology of neurodevelopmental disorders (for review see, e.g., ref. [30]). Our approach enabled the investigation of iPSC-derived cortical neurons during neurogenesis and to identify altered cell adhesion properties as well as impaired genesis of TBR1+ projection neurons in cells with ST3GAL3 deficiency.
In order to reduce genetic background effects, we used cells from a healthy sibling as control. The clones validated from patient and healthy sibling (ΔST3GAL3 and ASC23) did not show significant differences in their gene expression profiles. This indicates that DNA-methylation patterns of these cell lines did not differ much, suggesting that our control cell line fulfils the requirements expected for an isogenic control.
Pluripotency of iPSC lines was proved by the presence of LIN28A, NANOG, OCT4, PODXL, REX1, and SOX2 as markers. Moreover, gene expression patterns were comparable with human embryonic stem (hES) cells as shown by principal component analysis of RNA-sequencing data from iPSCs, hES, and feeder fibroblasts.
Further, the RNA-sequencing data showed only marginal differences between patient and control cells on iPSC level, while an independent cell line was clearly distinct. This is an additional indication that patient and control were genetically well matched. It also supports our hypothesis that the functional impairments leading to the observed phenotype in homozygous carriers of deleterious ST3GAL3 variants only come into full effect in differentiated cells.
RNA-sequencing of distinct human brain areas showed highest expression levels for ST3GAL3 in cortical samples, which is in line with the general consensus that the majority of epileptic spasms initiate from the human cortex and derive from an imbalance between excitatory and inhibitory signals [22]. Based on these findings, we used a protocol for cortical differentiation, which recapitulates the temporal and spacial pattern of in vivo neurogenesis [23]. This enabled us to investigate putative alterations in not only fully differentiated neuronal subtypes but also changes that can occur within a limited time window during neurogenesis.
Our first observation during these investigations was that ST3GAL3-deficient neuronal cells show a significantly increased speed of adherence after passaging as compared to controls. This is well in line with the known function of ST3GAL3, which is predominantly involved in forming sLea epitopes on proteins [10] known to be part of the ECM and/or the surface of the cell where they are crucially involved in the regulation of cell–cell and/or cell–matrix interactions (for review see, e.g., ref. [11]). Cell surface components as well as ECM components and composition have a strong influence on numerous functions and developmental processes in the nervous system, such as neural stem cell behaviour, cell migration and adhesion, synapse formation, etc. [31]. During cerebral development the ECM forms a basal lamina (BL), which modulates the maintenance of the neuronal stem cells and the differentiation, as well as migratory features of their descendants [32]. Interaction with the BL is thus vital for the survival of radial glia cells as well as cortical lamination [33]. The most important components of the BL are laminins, Tenascin-C, Reelin, and Notch-1 [34, 35]. Similar to Notch, which is sialylated, many of these adhesion molecules are glycosylated during cerebral development, and to date more than 70 genetic disorders based on disturbances in glycosylation are known (see, e.g., refs. [2, 36] for review). The most prominent symptoms of these CDGs are ID, epileptic seizures, and neuropathies (see, e.g., ref. [36]). With respect to sialylation defects in particular, it was found that St3gal2/3 double-knockout mice show impairment of motor coordination and of cognitive features [37]. Taken together, this underlines the importance of defects in protein glycosylation and sialylation in the aetiology of ID and related disorders and thus also points to a potential involvement of cellular adhesion features therein.
Still, the increased adherence velocity that we observed in cells from the affected individual can be explained in two ways: (i) by a shift in tissue composition towards cell types whose adherence behaviour is specifically affected and (ii) a general change affecting adherence features in all cell types. Moreover, a combination of both is possible.
However, because we found the number of TBR1-positive cells significantly reduced in samples from the affected individual, the first notion seems most likely. TBR1 is a marker for cortical projection neurons of lamina VI and belongs to a conserved family of genes whose products share the so-called T-box, a DNA-binding domain [38]. T-box transcription factors play a role in the regulation of developmental processes and were found to be critical for early murine cortical development. Specifically, Tbr1 controls both regional and laminar identity in postmitotic projection neurons [39].
Since the St3gal3 knockout mouse shows highly decreased levels of sialoglycoproteins but not in gangliosides, whole protein samples were targeted by lectin staining [37]. Staining of proteins derived from cortically differentiated cells with the lectin MAA revealed binding of the lectin to a band at about 80 kDa in control cells but reduced binding in ST3GAL3 mutant cells. Vice versa, the lectin DSA bound to a protein band at about 80 kDa in ST3GAL3 mutant cells, exclusively. DSA recognizes β-1,3-linked galactose residues (the target structure of ST3GAL3) and absence of ST3GAL3 was expected to cause a general increase of free β-1,3-linked galactose residues. However, the augmented detection of only one particular protein band by DSA in ST3GAL3 mutant cells suggests that ST3GAL3 might be highly substrate specific on the protein level. Thus, the identification of ST3GAL3 target proteins will be the focus of further analyses.
Taken together, our results corroborate the hypothesis that changes in the sialylation pattern on the surface of specific neuronal cell types affect adhesive interactions during development which may cause subtle changes in tissue composition, as also seen in patients suffering from glioneuronal malformations such as focal cortical dysplasias (see, e.g., ref. [40]), which share the occurrence of epilepsy as a common feature with the West syndrome phenotype of our patient. Moreover, these implications of ST3GAL3 in neural development may be relevant for the development and maintenance of normal cognitive functions.
References
Chiurazzi P, Pirozzi F. Advances in understanding - genetic basis of intellectual disability. F1000Res. 2016;5:F1000.
Hennet T, Cabalzar J. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction. Trends Biochem Sci. 2015;40:377–84.
Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49.
Chan B, Clasquin M, Smolen GA, Histen G, Powe J, Chen Y, et al. A mouse model of a human congenital disorder of glycosylation caused by loss of PMM2. Hum Mol Genet. 2016;25:2182–93.
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–63.
Hu H, Eggers K, Chen W, Garshasbi M, Motazacker MM, Wrogemann K, et al. ST3GAL3 mutations impair the development of higher cognitive functions. Am J Hum Genet. 2011;89:407–14.
Edvardson S, Baumann AM, Muhlenhoff M, Stephan O, Kuss AW, Shaag A, et al. West syndrome caused by ST3Gal-III deficiency. Epilepsia. 2013;54:e24–7.
Kato M. A new paradigm for West syndrome based on molecular and cell biology. Epilepsy Res. 2006;70(Suppl 1):S87–95.
Grahn A, Barkhordar GS, Larson G. Cloning and sequencing of nineteen transcript isoforms of the humanalpha2,3-sialyltransferase gene, ST3Gal III; its genomic organisation and expression in human tissues. Glycoconj J. 2002;19:197–210.
Ellies LG, Sperandio M, Underhill GH, Yousif J, Smith M, Priatel JJ, et al. Sialyltransferase specificity in selectin ligand formation. Blood. 2002;100:3618–25.
Kannagi R. Carbohydrate antigen sialyl Lewis a−its pathophysiological significance and induction mechanism in cancer progression. Chang Gung Med J. 2007;30:189–209.
Yamashita T, Hashiramoto A, Haluzik M, Mizukami H, Beck S, Norton A, et al. Enhanced insulin sensitivity in mice lacking ganglioside GM3. Proc Natl Acad Sci USA. 2003;100:3445–9.
Fragaki K, Ait-El-Mkadem S, Chaussenot A, Gire C, Mengual R, Bonesso L, et al. Refractory epilepsy and mitochondrial dysfunction due to GM3 synthase deficiency. Eur J Hum Genet. 2013;21:528–34.
Simpson MA, Cross H, Proukakis C, Priestman DA, Neville DC, Reinkensmeier G, et al. Infantile-onset symptomatic epilepsy syndrome caused by a homozygous loss-of-function mutation of GM3 synthase. Nat Genet. 2004;36:1225–9.
Thiesler CT, Cajic S, Hoffmann D, Thiel C, van Diepen L, Hennig R, et al. Glycomic characterization of induced pluripotent stem cells derived from a patient suffering from phosphomannomutase 2 congenital disorder of glycosylation (PMM2-CDG). Mol Cell Proteom. 2016;15:1435–52.
Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7:1836–46.
Wang Z, Ye S, Li J, Zheng B, Bao M, Ning G. Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol. 2011;11:109.
Konze SA, van Diepen L, Schroder A, Olmer R, Moller H, Pich A, et al. Cleavage of E-cadherin and beta-catenin by calpain affects Wnt signaling and spheroid formation in suspension cultures of human pluripotent stem cells. Mol Cell Proteom. 2014;13:990–1007.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19:782–9.
Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–16.
Bromfield EB, Cavazos JE, Sirven JI. An introduction to epilepsy. Bethesda: American Epilepsy Society: Distributed by NCBI; 2006. p. 1. electronic text
Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–86. S471
Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80.
Hevner RF. From radial glia to pyramidal-projection neuron: transcription factor cascades in cerebral cortex development. Mol Neurobiol. 2006;33:33–50.
Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–37.
Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–69.
Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–77.
Bajaj K, Madhusudhan MS, Adkar BV, Chakrabarti P, Ramakrishnan C, Sali, A, et al. Stereochemical criteria for prediction of the effects of proline mutations on protein stability. PLoS Comput Biol. 2007;3:e241.
Inoue H, Nagata N, Kurokawa H, Yamanaka S. iPS cells: a game changer for future medicine. EMBO J. 2014;33:409–17.
Barros CS, Franco SJ, Muller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol. 2011;3:a005108.
Porcionatto MA. The extracellular matrix provides directional cues for neuronal migration during cerebellar development. Braz J Med Biol Res. 2006;39:313–20.
Radakovits R, Barros CS, Belvindrah R, Patton B, Muller U. Regulation of radial glial survival by signals from the meninges. J Neurosci. 2009;29:7694–705.
Drago J, Nurcombe V, Bartlett PF. Laminin through its long arm E8 fragment promotes the proliferation and differentiation of murine neuroepithelial cells in vitro. Exp Cell Res. 1991;192:256–65.
Pierret C, Morrison JA, Rath P, Zigler RE, Engel LA, Fairchild CL, et al. Developmental cues and persistent neurogenic potential within an in vitro neural niche. BMC Dev Biol. 2010;10:5.
Freeze HH, Eklund EA, Ng BG, Patterson MC. Neurology of inherited glycosylation disorders. Lancet Neurol. 2012;11:453–66.
Yoo SW, Motari MG, Susuki K, Prendergast J, Mountney A, Hurtado A, et al. Sialylation regulates brain structure and function. FASEB J. 2015;29:3040–53.
Papaioannou VE. The T-box gene family: emerging roles in development, stem cells and cancer. Development. 2014;141:3819–33.
Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA. 2010;107:13129–34.
Becker AJ, Blumcke I, Urbach H, Hans V, Majores M. Molecular neuropathology of epilepsy-associated glioneuronal malformations. J Neuropathol Exp Neurol. 2006;65:99–108.
Acknowledgements
We thank Christian Sperling, Corinna Jensen, and Marcel Tauscher for excellent technical help and are grateful to Robert Weißmann for bioinformatic analyses. Further thanks is due to Herbert Hildebrandt for many helpful comments during creation of the manuscript. The machinery used during the course of this project was supported by a grant from the European Union (EFRE) to AWK. Researchers of Hannover Medical School were supported by funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) for the Cluster of Excellence REBIRTH (From Regenerative Biology to Reconstructive Therapy, EXC 62/2) and the SFB738, as well as the German Federal Ministry of Education and Research (PID-NET, 01GM1517F).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
41431_2018_220_MOESM1_ESM.docx
A patient-specific induced pluripotent stem cell model for West Syndrome caused by ST3GAL3 deficiency: Supplementary Information
Rights and permissions
About this article
Cite this article
van Diepen, L., Buettner, F.F.R., Hoffmann, D. et al. A patient-specific induced pluripotent stem cell model for West syndrome caused by ST3GAL3 deficiency. Eur J Hum Genet 26, 1773–1783 (2018). https://doi.org/10.1038/s41431-018-0220-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0220-5