Abstract
Bio-geographic ancestry is an area of considerable interest in the medical genetics, anthropology and forensics. Although genome-wide panels are ideal as they provide dense genotyping data, small sets of ancestry informative marker provide a cost-effective way to investigate genetic ancestry and population structure. Here, we investigate the performance of a reduced marker set that combine different types of autosomal markers through haplotype analysis. In particular, recently described DIP-STR markers should offer the advantage of comprising both, low mutation rate Indels (DIPs), to study human history over longer time scale; and high mutation rate STRs, to trace relatively recent demographic events. In this study, we assessed the ability of an initial set of 23 DIP-STRs to distinguish major population groups using the HGDP-CEPH reference samples. The results obtained applying the STRUCTURE algorithm show that the discrimination capacity of the DIP-STRs is comparable to currently used small-scale ancestry informative markers by approaching seven major demographic groups. Yet, the DIP-STRs show an improved success rate in assigning individuals to populations of Europe and Middle East. These data show a remarkable ability of a preliminary set of 23 DIP-STR markers to infer major biogeographic origins. A novel set of DIP-STRs preselected to contain ancestry information should lead to further improvements.
Similar content being viewed by others
Introduction
Bio-geographic ancestry inference has largely contributed to controlling for population structure in disease or trait association studies and to infer human evolutionary history [1,2,3,4]. Sets of small-scale ancestry informative markers (AIM) are valuable to provide most of the information in a cost-effective manner when sample sets have not been typed with genome-wide arrays of SNPs or when DNA amount is limited as in forensic science. Targeted association studies—such as candidate gene studies or replication studies following up genome-wide scans typically analyse a much smaller number of markers than genome-wide scans, making it difficult to infer ancestry in order to correct for stratification. AIMs are also used to select samples for follow-up studies before genome-wide scans are performed. In criminal investigations or missing persons identifications, in the absence of any other investigative leads (no database or suspect match), AIM genotypes obtained from evidential material could indicate the likely ancestry of the donor, and therefore help direct the course of investigations [5, 6].
Numerous AIM panels composed of dozens of autosomal SNPs, Indels or STRs have been published [7,8,9,10,11,12,13,14,15]. Briefly, these and other studies have shown that populations clustering patterns are robust, provided that at least about 60–150 markers are used [16,17,18], or about 30 or fewer if markers are preselected to have a high information content about ancestry [8, 12, 15, 19]. Most of small AIM sets are able to structure global populations into five major geographic regions: America, Sub-Saharan Africa, East Asia, Oceania and a cluster composed of Europe, Central-South Asia and Middle East populations within Eurasia [7].
Nevertheless, different types of markers offer different population structure resolution. In the field of molecular evolution, it has long been recognized that markers with relatively low mutation rates (SNP, Alu, Indel) serve as best loci for the analysis of human history over longer time scales (therefore to provide a biogeographical resolution at the level of continents), whereas rapidly evolving markers (STR and mtDNA) provide the greatest resolution over shorter time scales (regional resolution) [20]. Haplotypes composed of both slow and fast-evolving loci should combine the benefits of two types of markers. For example, in the case of linked SNP-STR, the instability of the STR should generate many alleles, in proportion to the populations’ divergence time, while the more stable flanking SNP should allow greater certainty in tracing the lineage of each haplotype. In addition, the number of possible haplotypes formed by the combination of these markers is much greater than the number of alleles at each individual locus. Such high variability increases the likelihood of rare haplotypes, that are easily lost during population founding and bottleneck events.
The first example of compound genetic markers are the SNP-STRs residing in the nonrecombining region of the human Y chromosome [20]. However, the uniparental transmission of the Y chromosome, like the mitochondrial genome, has the limit of reflecting only a fraction of individual’s ancestry with the risk of misinterpreting the overall ancestry in admixed individuals [6, 21]. To overcome such limitation Mountain et al. [22] proposed autosomal SNP-STR markers. In a pilot study they were able to show that two such compound markers were sufficient to provide support for the “out of Africa” human evolutionary hypothesis. Unfortunately, the application of SNP-STR for ancestry inference found limited success because of the difficulties initially encountered in genotyping SNPs linked STRs.
The current high throughput DNA sequencing technology finally enables genetic practitioners to consider multiple polymorphisms grouped into haplotypes. Many benchtop DNA sequencing platforms provide today continuous runs of a hundred base pairs or more on a single DNA molecule that allows to directly inferring the phase of the multiple markers within a small DNA segment. Minihaplotypes and microhaplotypes encompassing two to four SNPs spanning less than 10 kb and 200 bp, respectively, were recently described [23, 24]. Haplotype systems based on multiple SNPs have proven a forensically useful DNA marker for family or lineage inference [24, 25] and in anthropology for population relationships [19, 24, 26, 27]. A recent study showed that assignment of individuals to candidate populations significantly improves through combining linked SNPs. For example, incorrectly assigned individuals from empirical data of French and German populations can be decreased by 73% using haplotypes [28].
In this study, we evaluated the ability of an additional compound genetic marker recently described named DIP-STR, to serve as useful marker for ancestry inference. Originally, DIP-STRs were proposed as typing method for the characterization of DNA mixtures from two individuals present in very different proportions [29,30,31,32,33]. These mixtures are commonly encountered in forensic investigations during mixed trace analyses; in cases of peripheral blood DNA microchimerism during pregnancy or induced by solid organ transplant. Because of PCR amplification bias, the genetic identification of a DNA that contributes trace amounts to a mixed sample (minor DNA) represents a tremendous challenge. The DIP-STRs have the innovative feature of combining the analysis of a DIP (deletion/insertion polymorphism) and a closely linked STR polymorphism. To target the analysis of the minor DNA contributor of the mixture, PCR primers overlap the deleted/inserted sequence (S or L) to produce allele-specific amplifications of one or two minor DIP-STR haplotypes comprising the DIP allele that is not shared with the major DNA. Allele-specific amplifications of DIP-STR haplotypes enable the characterization a minor DNA in the presence of more than 1000-fold excess of a major DNA contributor.
Here we present the results of 23 DIP-STR markers typed in the global HGDP-CEPH reference samples. A comparative HGDP-CEPH analysis is shown using existing AIM SNP and AIM Indel marker sets.
Materials and methods
DIP-STR marker selection
DIP-STRs were selected from three groups: 14 from the first two sets of DIP-STRs developed for the analysis of unbalanced DNA mixtures (Table 1). Three markers from the first set were not included because of the large amplicon size (over 600 bp) and one appeared not interesting based on balanced allele frequencies of the DIP across the major geographic regions as reported by data from the 1000 Genomes project. Five additional DIP-STRs were rescued from the preliminary marker selection of the study [33]. In this previous study, several markers were initially eliminated because of the negative results of the first amplification assay; here they produced positive results with newly designed PCR primers. Among these candidates, we selected those markers with larger DIP allele frequencies differences between continents using the 1000 Genomes dataset. Finally, four DIP-STRs were selected considering previously described sets of AIM Indels [12,13,14, 34,35,36,37,38,39]. DIPs of interest were located near a repeated sequence, showed similar genotyping conditions and skewed global allele frequencies among continents.
Population samples
The CEPH Human Genome Diversity panel (CEPH-HGDP) contains 1,064 individuals from African, European, North African/Middle Eastern, Central-South Asian, East Asian, Native American and Oceanian populations [40]. For all data analyses purposes we considered only 952 individuals (H952 subset) after exclusion of duplicates, first and second-degree relatives [41]. Populations were combined into continental-based groups which have been previously established [16] with the following composite populations, sample sizes and labels: 6 African (105 AFR), 8 European (158 EUR), 4 North African/Middle Eastern (162 ME), 9 Central-South Asian (202 CSA), 17 East Asian (230 EAS), 2 Oceanian (28 OCE) and 5 Native American (64 NAM). For all subjects, blood cell samples were obtained according to protocols and informed-consent procedures approved by institutional review boards, and were labelled with an anonymous code number linked only to demographic information and sex.
Marker typing
DNA samples were genotyped for the 23 DIP-STRs and also for the 23 DIPs alone in order to further control for allele-specificity of the S and L-DIP-STR amplifications. PCR reactions were performed in 20 μL final volume. This contained 1 × PCR Buffer containing 1.5 mM MgCl2 (Thermo Fisher Écublens, Switzerland), 250 μM dNTP (Thermo Fisher Écublens, Switzerland), 1.2 U AmpliTaq Gold DNA Polymerase (Thermo Fisher Écublens, Switzerland) and 0.5 ng DNA. Primers’ sequences, quantities and multiplexes are indicated in Supplementary Table 1 and Supplementary Table 2. PCR thermal cycling conditions were: 5 min at 95 °C, 1 min at 94 °C, 1 min at annealing temperature specific to the markers set to be genotyped, 1 min at 72 °C for a number of PCR cycles that also varied across multiplex and a final extension of 30 min at 72 °C. Annealing temperatures and number of cycles are indicated in Supplementary Table 2.
PCR fragments were separated by capillary electrophoresis after adding 1 μL PCR amplicon to 8.5 μL deionized formamide HI-DI (Thermo Fisher Écublens, Switzerland) and to 0.5 μL 600 LIZ size standard (Thermo Fisher Écublens, Switzerland). Capillary electrophoresis was performed using an ABI PRISM 3130xl Genetic Analyzer (Thermo Fisher Écublens, Switzerland) according to the manufacturer’s instruction and analyzed using the GeneMapper® ID v3.2.1 software (Thermo Fisher Écublens, Switzerland), with a minimum peak height threshold of 50 RFU. The commercial DNA CEPH 1347–02 (Thermo Fisher Écublens, Switzerland) was added to two empty positions in each PCR plate as positive control of amplification and internal standard for allele calls, at least one empty well per plate was used a negative control of amplification. Markers information and genotypes are available at the HGDP-CEPH database (http://www.cephb.fr/en/hgdp_panel.php#basedonnees).
Data analysis
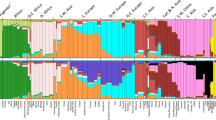
In order to assess the predictive value of the 23 DIP-STR marker set, cluster analysis was performed using the STRUCTURE program version 2.3.4 [42, 43]. Runs consisted of 50,000 Markov Chain steps after a burn-in of length 50,000 with ten replicates for K = 5 and K = 7, using the admixture ancestry model and correlated allele frequencies. The DIP-STR allele names indicated the DIP variant, either S (deletion) or L (insertion) and the STR allele expressed in DNA fragment size, this last corresponds to the STR allele size that one would obtain using primers located around the repeated sequence (allele names can be found in Fig. 1 and Supplementary Figure 1). For the STRUCTURE analysis DIP-STR alleles were recoded with serial numbers from 1 to n starting from the shortest L-DIP-STR to the longest S-DIP-STR. The bar plot was prepared using CLUMPAK (http://clumpak.tau.ac.il/).
To estimate the success rate in assigning the population of origin of each individual considering the genetic information of 23 DIP-STRs the likelihood-based approach implemented by Phillips et al. [44] in the SNIPPER App Suite (http://mathgene.usc.es/snipper/) was used. With this tool, a single, unknown genetic profile can be compared to a set of references populations, the “training set”. The software calculates individual maximum likelihoods estimates for the inclusion of the unknown sample into each reference population. A cross-validation has been performed using the frequency-based classifier in Snipper app suite. Each sample was tested in turn as unknown sample against the training set containing all the remaining samples. No marker deviated from the Hardy–Weinberg equilibrium and linkage disequilibrium. For comparative analysis, both STRUCTURE and Snipper-based analyses were repeated for two previously published AIM markers sets, these include a 34 AIM SNP set [44, 45] and a 46 AIM Indel set [37].
Results
Patterns of DIP-STR variability
The number of DIP-STR markers studied here is 23 (Table 1). Of the 368 alleles present more than once in the dataset, 27.5% appeared in all major regions represented, these are Africa, Europe, Middle East, Central-South Asia, East Asia, Oceania and Native America. Those exclusive to one region were 8.7%; region-specific alleles showed a median relative frequency of 1.4% in their region of occurrence.
As previously observed, Africa was the most variable region [46, 47]. Number of alleles, and mean number of private alleles followed a common trend: they were highest in the African samples, were somewhat lower in Europeans and East Asians, and were lowest in Amerindians and Oceanians. This was especially true for 13 markers (Supplementary Figure 1). Note that Oceanians population size is about half the Native Americans and between one fourth and one eight of the other major geographic regions. In Fig. 1a is showed the allele frequency distribution of marker MID1013-D5S490 representative of the pattern described above. Conversely, seven markers showed a higher number of alleles outside Africa. For these markers the DIP variant is not polymorphic in Africa therefore all the DIP-STR haplotypes containing the DIP allele that probably appeared outside Africa, are missing in this group. Here as well, the number of observed alleles decreases with increasing distance from Eurasia. An example is reported in Fig. 1b, marker rs112604544-STR.
Population clustering analysis
We assessed the ability of the DIP-STR genotypes to cluster the global HGDP-CEPH reference population samples applying the widely used STRUCTURE Bayesian grouping algorithm. Previous studies including several types of ancestry informative markers suggest that when limited numbers of markers are analyzed it is appropriate to aim to assign individuals to five major population groups in the first instance, these are Africans, Europeans, East Asians, Oceanians and Native Americans. To obtain the clearest pattern of group membership from STRUCTURE, the study population complexity was reduced by excluding geographically close populations such as Middle East and Central-South Asia. These are known to show higher misclassification rates since they occupy regions in the middle of a continuum of variability. The STRUCTURE results with the highest likelihood at K = 5 shows a clear pattern of five clusters corresponding to major geographical regions (Fig. 2a). These results are comparable to currently used AIM markers sets comprising 34 AIM SNPs (Fig. 2b) or 46 AIM Indels (Fig. 2c). The results at K = 7 including the complete HGDP-CEPH dataset show that although a “private” ancestry component is present in Middle East and Central South Asia, clustering patterns are less distinct and Europe loses clear definition incorporating the additional inferred cluster as partial degrees of ancestry (Fig. 3a). These data are comparable to the results obtained using the 34 AIM SNP set (Fig. 3b), yet they show an improved clustering capacity of the European and Middle East groups when compared to the results obtained with the 46 AIM Indels (Fig. 3c).
Individual ancestry assignment analysis
Snipper cross-validation classification success values for five and seven HGDP-CEPH reference samples obtained using the 23 DIP-STRs considered in this study are in agreement with the clustering obtained using the STRUCTURE classification algorithm. For the five group analysis the classification success rates are higher than 99% for all populations. These results are similar to those obtained with 34 AIM SNPs and 46 AIM Indels, with a somewhat better classification success rate for the East Asian group (Table 2). For the seven group analysis, samples with Eurasians origin were more difficult to classify, yet about 96% of Europeans 86% of Middle Easterns are correctly classified (Table 3). These values are lower when using 34 AIM SNPs (70 and 62%) and 46 AIM Indels (40 and 46%).
Separate STRUCTURE analysis of the 23 DIP set and the 23 STR set
The independent contribution to the clustering of the HGDP-CEPH populations of the 23 DIPs and the 23 STRs composing the haplotypes analyzed before, was investigated (Fig. 4). This is possible because haplotypes are named based on the respective DIP and STR comprising alleles. The data at K = 5 after excluding geographically close populations of Middle East and Central South Asia, show similar results for the DIPs and the STRs. The populations of Africa are distinguished from the other main geographical regions while part of Native Americans are clustered with the Oceanians. The populations of Eurasia form a unique cluster with a high degree of noise and some indication of admixture with the Native Americans, especially when using the DIPs.
Discussion
The aim of this study was to explore the contribution to small-scale marker sets based biogeographic inference of DIP-STR haplotype markers. We hypothesized that the compound nature of DIP-STRs may represent an attractive feature not only for DNA mixture resolutions as we originally proposed, but also for studies of population structure. Briefly, each DIP-STR haplotype is provided with a slow and fast mutating variant that should confer higher biogeographic information compared to the use of each single type of polymorphism. Here, the survey of the HGDP-CEPH global reference samples with an initial set of 23 DIP-STRs allowed us to determine the relative value of our markers with respect to other validated AIM marker sets.
Overall, the clustering patterns observed with the STRUCTURE algorithm and the population assignments obtained with the Snipper program were in good correlation with each other as well as with worldwide population structure [8, 12, 15, 19, 37]. The prediction of bio-geographic ancestry was achieved for five major populations using a smaller set of markers (23 DIP-STRs vs. 34 to 46 markers of validated AIM SNP and AIM Indel marker sets). Moreover, the seven group analysis shows that with these markers the global pattern is approaching seven clusters and some additional selected DIP-STRs may produce stable and reproducible distinction of the populations of Middle East and Central-South Asia within Eurasia. The discrimination of these groups still represents a challenge for most available AIM panels [7], as also indicated by our comparative analysis using 34 AIM SNPs and 46 AIM Indels. However, according to Snipper cross-validation success values the DIP-STRs show an improved success rate in assigning individuals to populations of Europe and Middle East. It should be noted that the results showed here, were obtained with DIP-STR markers not selected for distinguishing continental or intra-continental structures. Yet, these results are comparable to those obtained with larger sets of SNPs and Indels produced after refined selections using allele frequency data from worldwide population surveys. Finally, the fact that the obtained resolution is not due to single composing markers (either DIPs or STRs) with no advantage of considering the combined information, is also showed by the corresponding cluster analysis using the DIP marker set and the STR marker set, separately.
It is generally accepted that identifying SNPs that can distinguish among four or five continental groups of populations is not difficult [48]. Many different non-overlapping sets of SNPs capable of inferring continental ancestry were described (see references in the Introduction). However, previous studies showed the difficulty of developing a single panel comprising a limited number of SNPs or Indels that is capable of differentiating populations both globally and within regions. Note that, the HGDP collection has the drawback of not sampling densely within each geographic region; however, the use of a large number of markers showed the possibility of a sub-continental estimation of bio-geographic ancestry.
To reach such a result with DIP-STR markers; first of all, they need to be selected for ancestry purposes. Large whole genome sequencing data will provide the basis for annotating novel DIP-STRs and estimating allele frequencies across populations from long read data; alternatively, additional marker selecting criteria can be formulated based on annotated lists of unphased DIP and STR markers. For example: a tentative approach could be a marker selection based on DIP skewed allele frequencies among large geographical regions (this would include a mixture of old and young DIP mutations) linked to highly polymorphic STRs.
Finally, the value of a small ancestry panel is also measured based on the capacity of analyzing trace amount of DNA with a cost-effective method easy to implement in routine laboratories. Most of the DIP-STRs genotyped here were analyzed in short fragments by standard PCR and capillary electrophoresis analysis and were validated for forensic contact traces [32]. Yet, several multiplex were used to produce the data. This is because: on one side, markers were progressively added to the study; on the other side, the typing of 23 DIP-STRs conserving S and the L-specific amplifications require some developmental efforts. We plan to work on a compact multiplex method once final DIP-STRs and minimum number of marker are identified.
As stated before, significant progress has been made in the research of ancestry marker sets; therefore the effort that the development of AIM-DIP-STRs requires is justified only by an advantage over existing methods. Besides the hope of providing better (more refined and robust) resolution of bio-geographic ancestry that can be assessed with simple and cost-effective methods, these markers have the attractive feature of combining ancestry estimates and unbalanced DNA mixture resolution, especially useful in forensics. Although, ancestry and identity DIP-STRs are may not to be the same marker sets, one can apply DIP-STR markers to first, detect the minor DNA of the trace sample; for example, the DNA of a man who sexually assaulted a woman, retrieved from a gynecological sample. Second, if the DIP-STR profile does not find a match in the list of suspects, additional ancestry DIP-STRs can be used as intelligence tool to further guide the investigation. A caveat to this is that the ancestry inference in the context of mixture resolution would produce results for a reduced number of markers (those that contain DIP alleles unique to the minor DNA) and therefore the validation of redundant DIP-STRs may be required to have sufficient markers for ancestry resolution.
Our study provides another example of forensic markers capable of providing substantial biogeographic information. The fact that individual identifiability and population identifiability may be correlated has been observed before for forensic microsatellite markers [49] and it has been extensively studied by Algee-Hewitt and colleagues [50]. These authors identified in the high level of polymorphism the potential for high population identifiability. In the case of DIP-STRs the number of possible haplotypes formed by the combination of two markers is indeed much greater than the number of alleles at each individual locus, but the combination of slow- and fast- evolving loci is also expected to play a role.
In conclusion, our study demonstrates that DIP-STRs can provide a useful adjunct to AIM panels aiming at providing a better resolution and, in forensic science, at combining the function of mixture deconvolution to bio-geographic ancestry estimates. We showed here that continental regions can be readily distinguished, while more markers are necessary to improve the classification of closely related populations such as Eurasians.
This represents an exceptional result for the first evaluation of this set of marker that was not selected to contain information about ancestry. The research described here should be considered a pilot study to encourage more efforts on DIP-STRs marker discovery and multiplex development. Finally, these data provide references of global DIP-STR allele frequencies including the set proposed for forensic casework analysis.
References
Hellenthal G, Busby GB, Band G, et al. A genetic atlas of human admixture history. Science. 2014;343:747–51.
Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4.
Tishkoff SA, Kidd KK. Implications of biogeography of human populations for “race” and medicine. Nat Genet. 2004;36:S21–7.
Elhaik E, Tatarinova T, Chebotarev D, et al. Geographic population structure analysis of worldwide human populations infers their biogeographical origins. Nat Commun. 2014;5:3513.
Kayser M, de Knijff P. Improving human forensics through advances in genetics, genomics and molecular biology. Nat Rev Genet. 2011;12:179–92.
Phillips C, Prieto L, Fondevila M, et al. Ancestry analysis in the 11-M Madrid bomb attack investigation. PLoS ONE. 2009;4:e6583.
Phillips C. Forensic genetic analysis of bio-geographical ancestry. Forensic Sci Int Genet. 2015;18:49–65.
de la Puente M, Santos C, Fondevila M, et al. The global AIMs nano set: a 31-plex SNaPshot assay of ancestry-informative SNPs. Forensic Sci Int Genet. 2016;22:81–8.
Pakstis AJ, Kang L, Liu L, et al. Increasing the reference populations for the 55 AISNP panel: the need and benefits. Int J Legal Med. 2017;131:913–7.
Phillips C, Parson W, Lundsberg B, et al. Building a forensic ancestry panel from the ground up: the EUROFORGEN Global AIM-SNP set. Forensic Sci Int Genet. 2014;11:13–25.
Lao O, van Duijn K, Kersbergen P, de Knijff P, Kayser M. Proportioning whole-genome single-nucleotide-polymorphism diversity for the identification of geographic population structure and genetic ancestry. Am J Hum Genet. 2006;78:680–90.
Santos C, Phillips C, Oldoni F, et al. Completion of a worldwide reference panel of samples for an ancestry informative Indel assay. Forensic Sci Int Genet. 2015;17:75–80.
Zaumsegel D, Rothschild MA, Schneider PM. A 21 marker insertion deletion polymorphism panel to study biogeographic ancestry. Forensic Sci Int Genet. 2013;7:305–12.
Yang N, Li HZ, Criswell LA, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118:382–92.
Londin ER, Keller MA, Maista C, et al. CoAIMs: a cost-effective panel of ancestry informative markers for determining continental origins. PLoS ONE. 2010;5:e13443.
Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–5.
Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: Utility and applications. Hum Mutat. 2008;29:648–58.
Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78.
Kidd KK, Pakstis AJ, Speed WC, et al. Current sequencing technology makes microhaplotypes a powerful new type of genetic marker for forensics. Forensic Sci Int Genet. 2014;12:215–24.
de Knijff P. Messages through bottlenecks: On the combined use of slow and fast evolving polymorphic markers on the human Y chromosome. Am J Hum Genet. 2000;67:1055–61.
King TE, Parkin EJ, Swinfield G, et al. Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy. Eur J Hum Genet. 2007;15:288–93.
Mountain JL, Knight A, Jobin M, et al. SNPSTRs: Empirically derived, rapidly typed, autosomal Haplotypes for inference of population history and mutational processes. Genome Res. 2002;12:1766–72.
Kidd KK, Speed WC, Pakstis AJ, et al. Evaluating 130 microhaplotypes across a global set of 83 populations. Forensic Sci Int Genet. 2017;29:29–37.
Pakstis AJ, Fang R, Furtado MR, Kidd JR, Kidd KK. Mini-haplotypes as lineage informative SNPs and ancestry inference SNPs. Eur J Hum Genet. 2012;20:1148–54.
Kidd KK, Pakstis AJ, Speed WC, et al. Microhaplotype loci are a powerful new type of forensic marker. Forensic Sci Int Genet Suppl Ser. 2013;4:e123–4.
Kidd JR, Friedlaender F, Pakstis AJ, et al. Single nucleotide polymorphisms and haplotypes in Native American populations. Am J Phys Anthropol. 2011;146:495–502.
Schlebusch CM, Soodyall H. Extensive population structure in San, Khoe, and mixed ancestry populations from southern Africa revealed by 44 short 5-SNP haplotypes. Hum Biol. 2012;84:695–724.
Gattepaille LM, Jakobsson M. Combining markers into haplotypes can improve population structure inference. Genetics. 2012;190:159–74.
Castella V, Gervaix J, Hall D. DIP-STR: highly sensitive markers for the analysis of unbalanced genomic mixtures. Hum Mutat. 2013;34:644–54.
Cereda G, Biedermann A, Hall D, Taroni F. An investigation of the potential of DIP-STR markers for DNA mixture analyses. Forensic Sci Int Genet. 2014;11:229–40.
Cereda G, Biedermann A, Hall D, Taroni F. Object-oriented Bayesian networks for evaluating DIP-STR profiling results from unbalanced DNA mixtures. Forensic Sci Int Genet. 2014;8:159–69.
Oldoni F, Castella V, Grosjean F, Hall D. Sensitive DIP-STR markers for the analysis of unbalanced mixtures from “touch” DNA samples. Forensic Sci Int Genet. 2017;28:111–7.
Oldoni F, Castella V, Hall D. A novel set of DIP-STR markers for improved analysis of challenging DNA mixtures. Forensic Sci Int Genet. 2015;19:156–64.
Bastos-Rodrigues L, Pimenta JR, Pena SDJ. The genetic structure of human populations studied through short insertion-deletion polymorphisms. Ann Hum Genet. 2006;70:658–65.
Francez PA, Ribeiro-Rodrigues EM, dos Santos SE. Allelic frequencies and statistical data obtained from 48 AIM INDEL loci in an admixed population from the Brazilian Amazon. Forensic Sci Int Genet. 2012;6:132–5.
Pereira R, Phillips C, Alves C, Amorim A, Carracedo A, Gusmao L. A new multiplex for human identification using insertion/deletion polymorphisms. Electrophoresis. 2009;30:3682–90.
Pereira R, Phillips C, Pinto N, et al. Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS ONE. 2012;7:e29684.
Rosenberg NA, Mahajan S, Ramachandran S, Zhao CF, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:660–71.
Santos NPC, Ribeiro-Rodrigues EM, Ribeiro-dos-Santos AKC, et al. Assessing individual interethnic admixture and population substructure using a 48-Insertion-Deletion (INSEL) Ancestry-Informative Marker (AIM) Panel. Hum Mutat. 2010;31:184–90.
Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–2.
Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–7.
Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87.
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59.
Phillips C, Salas A, Sanchez JJ, et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int-Genet. 2007;1:273–80.
Fondevila M, Phillips C, Santos C, et al. Revision of the SNPforID 34-plex forensic ancestry test: assay enhancements, standard reference sample genotypes and extended population studies. Forensic Sci Int Genet. 2013;7:63–74.
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–7.
Calafell F, Shuster A, Speed WC, Kidd JR, Kidd KK. Short tandem repeat polymorphism evolution in humans. Eur J Hum Genet. 1998;6:38–49.
Soundararajan U, Yun L, Shi M, Kidd KK. Minimal SNP overlap among multiple panels of ancestry informative markers argues for more international collaboration. Forensic Sci Int Genet. 2016;23:25–32.
Phillips C, Fernandez-Formoso L, Garcia-Magarinos M, et al. Analysis of global variability in 15 established and 5 new European Standard Set (ESS) STRs using the CEPH human genome diversity panel. Forensic Sci Int Genet. 2011;5:155–69.
Algee-Hewitt BF, Edge MD, Kim J, Li JZ, Rosenberg NA. Individual identifiability predicts population identifiability in forensic microsatellite markers. Curr Biol. 2016;26:935–42.
Acknowledgements
This work was financially supported by funding from the University Center of Legal Medicine of the University Hospital of Lausanne and the Faculty of Biology and Medicine of the University of Lausanne (Pro-Femmes fellowship to Diana Hall).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Moriot, A., Santos, C., Freire-Aradas, A. et al. Inferring biogeographic ancestry with compound markers of slow and fast evolving polymorphisms. Eur J Hum Genet 26, 1697–1707 (2018). https://doi.org/10.1038/s41431-018-0215-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0215-2
This article is cited by
-
Early noninvasive prenatal paternity testing by targeted fetal DNA analysis
Scientific Reports (2023)