Abstract
Alzheimer’s disease (AD) and frontotemporal dementia (FTD) are the two most common neurodegenerative dementias. Variants in APP, PSEN1 and PSEN2 are typically linked to early-onset AD, and several genetic risk loci are associated with late-onset AD. Inherited FTD can be caused by hexanucleotide expansions in C9orf72, or variants in GRN, MAPT or CHMP2B. Several other genes have also been linked to FTD or FTD with motor neuron disease. Here we describe a cohort of 60 Finnish families with possible inherited dementia. Our aim was to clarify the genetic background of dementia in this cohort by analysing both known dementia-associated genes (APOE, APP, C9ORF72, GRN, PSEN1 and PSEN2) and searching for rare or novel segregating variants with exome sequencing. C9orf72 repeat expansions were detected in 12 (20%) of the 60 families, including, in addition to FTD, a family with neuropathologically verified AD. Twelve families (10 with AD and 2 with FTD) with representative samples from affected and unaffected subjects and without C9orf72 expansions were selected for whole-exome sequencing. Exome sequencing did not reveal any variants that could be regarded unequivocally causative, but revealed potentially damaging variants in UNC13C and MARCH4.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) and frontotemporal dementia (FTD) are the two most common neurodegenerative dementias. AD is characterized by progressive loss of memory, typically presenting with deficits in anterograde episodic memory. Other cognitive functions, such as language, executive functions and visuospatial functions, deteriorate as the disease progresses [1]. Most AD patients first develop symptoms after 65 years of age (late-onset AD, LOAD), while <10% of patients present with early-onset AD (EOAD). Autosomal dominant inheritance and rare cases of autosomal recessive inheritance are seen in the EOAD group, due to variants in the amyloid precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2) genes. Familial cases are also seen in LOAD with an estimated heritability of 58–79% [2]. More than 20 disease-associated loci have been detected in genome-wide association studies (GWAS) [3,4,5,6,7] and meta-analyses [8], but apart from the APOE ε4 and TREM2 p.(Arg47His) risk alleles, most of these only have a modest effect. Although rare variants in APP, PSEN1 and PSEN2 have been detected in LOAD patients by targeted resequencing [9], variants affecting function are rare in LOAD and its pathobiology reflects the interplay of predisposing genetic variants and environmental factors. Sequencing studies have also shown that rare variants can be found in dementia-associated loci identified through GWAS [10].
In contrast to AD, FTD is more commonly observed in patients younger than 65 years [11]. FTD may present with changes in personality and behaviour (behavioural-variant FTD) or language difficulties (non-fluent variant primary progressive aphasia and semantic-variant progressive aphasia) [11]. Up to 40% of patients have a positive family history with autosomal dominant inheritance in 10% [12]. The most commonly mutated genes are C9orf72 [13, 14], GRN [15, 16] and MAPT [17], while rare variants in TARDBP [18], FUS [19, 20], VCP [21], CHMP2B [22], UBQLN2 [23, 24], TBK1 [25], SQSTM1 [26] and CCNF [27] have been detected in patients with either FTD and motor neuron disease (FTD-MND) or pure FTD. C9orf72 expansions are prevalent in Finnish patients with FTD or ALS, accounting for 48 and 46% of familial FTD and ALS, respectively, and 19 and 21% of sporadic FTD and amyotrophic lateral sclerosis (ALS) [13].
Based on the known functions of disease-associated genes, several pathways involved in the pathogenesis of AD and FTD have been identified. In AD, these include the amyloid β pathway, the immune system (CLU, CR1, ABCA7, CD33, EPHA1, the MS4A gene cluster), synaptic activity (PICALM, CD33, CD2AP, EPHA1, BIN1) and lipid metabolism (CLU, ABCA7) (reviewed in ref. [28]). In FTD, the disease-implicated pathways include RNA processing and transcription regulation (C9orf72, TARDBP, FUS), microtubule function (MAPT), immune response (GRN), lysosome-mediated and ubiquitin-mediated protein degradation and autophagy (GRN, VCP, CHMP2B) (reviewed in ref. [11]).
Here we describe a cohort of 60 Finnish families with possible inherited dementia. Our aim was to clarify the genetic background of dementia in this cohort by analysing both known dementia-associated genes and searching for rare or novel segregating variants using exome sequencing. We show that C9orf72 hexanucleotide repeat expansions are common in this cohort but variants affecting function of the other most common AD and FTD genes are not accountable for the disease in these families. We also present rare variants that segregate with AD and FTD in small families. Although no definite conclusion can be achieved regarding the causal involvement of these rare variants, these should be taken into account in future studies trying to identify the genetic cause of familial dementias.
Subjects and methods
Study cohort
The study cohort is comprised of affected and unaffected members from 60 Finnish families with possible inherited dementia. The families were recruited from neurology clinics in the Helsinki and Uusimaa hospital district (Southern Finland) and via an advertisement in a national newspaper in the late 1990s. The recruitment method proved particularly successful, resulting in 60 families suited for the study. A total of 364 blood-derived DNA samples (107 from affected patients and 257 from unaffected family members) were available from the families. A prerequisite for participation was a positive family history with two or more living first-degree family members affected by dementia.
The clinical diagnosis was AD in most families (n = 38), FTD in 10 families, dementia with Lewy bodies (DLB) in one family and unspecified dementia in 11 families (Supplementary table 1). The diagnoses were based on clinical findings and brain computed tomography (CT) imaging studies. Extensive neuropsychological studies had been performed for some of the patients. Liquor biomarkers were not available at the time of patient recruitment and sample collection. Neuropathological data was available from patients belonging to seven of the 60 families. The ages at onset are listed in Supplementary table 2.
Ethical aspects
Informed consent was obtained from all participants. The study was approved by the Ethics committee of Neurology department at HUCH (4.6.1997 and 11.1.2012) and the HUCH Ethics Committee of Medicine (Dnro104/13/03/01/14). Approval for using patient tissue specimens was given by Valvira (Dnro 2855/06.01.03.01/2012). Approval for using medical records and autopsy reports of the patients living outside the HUS district was obtained from National Institute for Health and Welfare (Dnro THL/701/5.05.00/2013).
Methods
EDTA blood samples were drawn after obtaining informed consent from the participants. Both affected patients and unaffected family members were recruited for the study. DNA was extracted using standard protocols. Overview of the study scheme is shown in Fig. 1. APOE genotypes were determined by PCR and CfoI digestion as described in Myllykangas et al. [29]. Screening of C9orf72 expansions was done by repeat-primed PCR as described by Renton et al. [13].
Expansion was defined by two criteria that had to be fulfilled: (1) characteristic saw-tooth pattern in repeat-primed PCR extending over 30 G4C2 repeats on capillary electrophoresis of the PCR products, and (2) lack of large allele (>30 repeats) amplicon in standard PCR across the repeat region. Standard PCR across the repeat region was performed using LongAmp Taq Reaction Buffer (New England Biolabs, Ipswich, MA, USA) with the following PCR primers 5′-GGAGGGAAACAACCGCAGCCTGTAG-3′ and 5′-ATGCCGCCTCCTCACTCACCCACTCG-3′, 1.8 M of Betaine. The PCR products were run on 2% agarose gels.
We evaluated the pedigrees for the availability of samples from both affected and unaffected individuals as well as the availability of neuropathological data. Twelve families without C9orf72 expansions (2 with FTD and 10 with AD) were selected for further genetic studies. The FTD families were screened for variants in GRN and the AD families for variants in exons 16 and 17 of APP and the coding regions of PSEN1 and PSEN2. Exons and flanking splice site regions were amplified by PCR and the purified PCR products were sequenced in both directions using the BigDye Terminator v3.1 cycle sequencing Kit (Applied Biosystems, CA, USA). All primer sequences and PCR conditions are available upon request.
Large structural and copy-number variants were excluded by using HumanOmniExpress Bead Chip (Illumina, San Diego, CA, USA). Loci known to have copy-number variants that associate with dementia (such as APP and SNCA) were checked visually. In addition, the data was analysed with CNVPartition in Genome Studio (Illumina, San Diego, CA, USA) to detect large (>50 kb) CNVs. The identified CNVs were checked against the Database of Genomic Variants [30].
Whole-exome sequencing (WES) of selected individuals was done at University College London (UCL, London, UK). Exome enrichment was performed using TruSeq Exome Capture kit (Illumina, San Diego, CA, USA). Sequencing was performed on a HiSeq 2000 (Illumina). Reads were aligned to GRCh37/hg19 using BWA, variants called according to GATK best practice guidelines and annotated with ANNOVAR [31]. In silico pathogenicity predictions of non-synonymous variants were done with SIFT [32], Polyphen2 [33], MutationTaster [34], MutationAssessor [35] and CADD [36]. Variants were filtered against population databases (1000Genomes, ESP and ExAC) and prioritized based on variant type (missense, nonsense, splice site, frameshift, non-frameshift) and predicted pathogenicity. We concentrated on variants found in genes implicated in GWAS or genes that are highly expressed in the brain. We also assessed the known functions of the genes of interest.
Selected variants (shared by the affected members in each family but not present in the unaffected family member, if appropriate sample was available) from WES were confirmed with Sanger sequencing and their segregation tested in a family setting. These variants and the associated phenotypes have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) with accession numbers SCV000576395, SCV000576396, SCV000576397, SCV000576398 and SCV000576399. We also checked the frequencies of these selected variants in SISu, a database of sequence variants in Finns (Sequencing Initiative Suomi project (SISu), Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Finland (http://sisuproject.fi), SISu v4.1, accessed in 09/2016).
Results
APOE genotyping
APOE genotyping was performed for 364 samples from 60 families. There were 34 individuals homozygous for the risk APOE genotype ɛ4 (20/107 affected, 18.7%; 14/257 unaffected, 5.4%). A total of 166 individuals were heterozygotes APOE ɛ3/ɛ4 (56/107 affected, 52.3%; 110/257 unaffected, 42.8%). The most common allele, APOE ɛ3, is not associated with an increased risk for AD, and it was detected in 28 of 107 (26.1%) affected patients and in 123 of 257 (47.8%) unaffected individuals. Five unaffected individuals (5/257, 1.99%) were APOE ɛ2/ɛ4 heterozygotes and one unaffected (1/257, 0.3%) was ɛ2/ɛ3. No genotype was obtained for seven samples (three affected and four unaffected). The genotypes in each family are shown in Supplementary table 2.
C9orf72
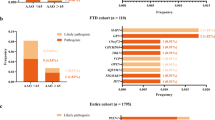
All 60 families were included in the C9orf72 hexanucleotide expansion screening. Expansions were detected in 12 of the 60 families (20%). The distribution of expansions in affected and unaffected individuals in each family is shown in Supplementary table 2. Clinical diagnosis was FTD in seven families, AD or variant AD in four families and degenerative dementia (ALS, DLB or AD-resembling syndromes) in one family (Table 1). The proportions of C9orf72 expansions in each diagnosis group is shown in Fig. 2. As FTD or FTD/ALS are the typical clinical phenotypes associated with C9orf72 expansions, we only describe the five families with more atypical presentations in detail. No additional information was available from family Fam-62.
The index patient of family Fam-18 developed symptoms at 65 years. Clinical presentation was compatible with AD, but CT and magnetic resonance imaging studies showed frontal atrophy. Clinical data from other family members was not available.
Patient records of two affected patients from family Fam-27 were available for review. Both patients had clinical AD with late onset.
Neuropathological data of two affected patients from family Fam-31 were available. The index patient had LOAD and the diagnosis was verified neuropathologically post mortem in 1998 using the methods available at that time. Re-analysis of the archived formalin-fixed, paraffin-embedded sample revealed Braak stage V tau-pathology and CERAD stage B beta-amyloid load. TDP-43 staining was negative. p62-positive inclusions were observed in the granular cerebellar cells. No DNA sample was available for study. Formalin-fixed brain tissue from temporal lobe of another patient of the family was available for immunohistochemistry. TDP-43 staining was negative, but moderate to severe tau-pathology suggestive of AD was observed. This patient was shown to harbour a C9orf72 expansion and carried the APOE ɛ4/4 genotype.
Family Fam-50 included several affected individuals with onset of disease after 70 years of age. DNA sample was available from one of them. One patient had visual hallucinations as the first symptom and subsequently developed loss of concentration and memory deficit. Neuropsychological examination was consistent with large-scale impairment and visual defect.
The index patient of family Fam-73 was diagnosed with variant AD. However, brain SPECT was suspective of FTD. The index patient’s sister had been diagnosed with ALS. Thus, the actual diagnostic spectrum of this family is consistent with FTD and ALS.
Further studies on 12 families without C9orf72 expansions
Exclusion of GRN, APP, PSEN1 and PSEN2
Twelve families without C9orf72 expansions and representative samples from both affected and unaffected individuals were selected for further studies (Fig. 1.). The clinical diagnoses in these families were FTD (two families: Fam-13 and Fam-59) and AD (10 families: Fam-15, Fam-29, Fam-32, Fam-35, Fam-38, Fam-49, Fam-52, Fam-55, Fam-56 and Fam-57). Sanger sequencing did not reveal any causal variants in GRN (FTD families), APP exons 16 and 17, or coding regions of PSEN1 and PSEN2 (AD families).
Exclusion of large structural and copy-number variants by SNP microarray
None of the 12 index patients had a duplication of APP. Neither did we identify any deletions or duplications involving other known dementia-associated genes, such as SNCA.
Whole-exome sequencing
WES data was generated for at least two affected patients from each of the 12 families. The oldest unaffected family members from whom a DNA sample was available (seven families) were exome sequenced as controls. We concentrated on rare variants identified in WES shared by the affected patients in each family but not seen in the analysed healthy family members, when available (list of rare variants in each family can be found in Supplementary table 3). Confirmation and segregation analyses were done with Sanger sequencing. A large number of shared rare variants were identified in each family, but we concentrated on variants in GWAS hit genes or in genes with known functions possibly relevant for neurodegeneration. The validated variants are listed in Table 2 and presented in detail below.
WES findings in AD families
CLU
A heterozygous CLU c.608C>T, p.(Thr203Ile) variant (rs41276297) was identified in two affected patients of family Fam-56. The variant was not detected in the four unaffected family members from whom a sample was available (Fig. 3a). This variant is a previously known, rare variant with a frequency of 0.00121 in Finnish samples in ExAC. Polyphen and SIFT predicted no deleterious effect. This variant has also been detected in British AD samples (reported as p.T255I) with a frequency of 0.003 as well as in unaffected controls (frequency 0.006) [37]. One of the affected individuals also carried one APOE ɛ4 allele, while the other was homozygous for ɛ3.
Pedigrees of the families with rare variants verified by Sanger sequencing. DNA samples were available from individuals marked with an asterisk. APOE genotypes are also marked in the pedigree. a Family Fam-56 with the CLU p.(Thr203Ile) variant. Heterozygous variant (−/+), homozygous wild-type allele (−/−). b Family Fam-15 with the PCDH11X p.(Asp760Val) variant. Heterozygous variant (−/+), homozygous wild-type allele (−/−), hemizygous variant (+), hemizygous wild-type allele (−). c Family Fam-49 with the UNC13C p.(Lys443del) variant. Heterozygous variant (−/+), homozygous wild-type allele (−/−). d. Family Fam-13 with the MARCH4 p.(Lys211Glu) variant. Heterozygous variant (−/+), homozygous wild-type allele (−/−). e Family Fam-59 with the MARCH4 p.(Trp13Cys) variant. Heterozygous variant (−/+), homozygous wild-type allele (−/−)
PCDH11X
Affected members of family Fam-15 carried a heterozygous c.2279A>T, p.(Asp760Val) variant in PCDH11X. This variant (rs781770086) is present as a singleton in ExAC (a potential low-quality site) and in SISu. Sanger sequencing confirmed the variant in the two affected patients (II:4 and III:4). However, segregation analysis showed that the variant was also present in two currently unaffected individuals (III:2 and III:5) and in one individual (III: 6) with unclear status. The remaining two unaffected family members (II:7 and III:7) did not carry the variant (Fig. 3b). Only one of the affected individuals carried APOE ɛ4.
UNC13C
In family Fam-49, a heterozygous 3-bp deletion in UNC13C, c.1324_1326del, p.(Lys443del) was detected in two affected patients. The variant was not seen in two unaffected family members (Fig. 3c). This in-frame deletion variant (rs746069739) is present as a singleton in ExAC and in SISu (a low-quality site). In addition to the UNC13C variant, both affected individuals also carried one APOE ɛ4 allele.
WES findings in FTD families
MARCH4
Two affected patients from the FTD family Fam-13 carried a heterozygous c.631A>G, p.(Lys211Glu) variant, (rs756981946) in MARCH4. This variant was absent from the unaffected family members (Fig. 3d). The APOE genotypes of the two affected individuals of Fam-13 were ɛ3/4 and ɛ3/3. Two affected members of the second FTD family, Fam-59, carried a heterozygous c.39C>G variant, p.(Trp13Cys) (rs145386484) in MARCH4. Segregation analysis showed that this variant was also present in seven currently unaffected family members (ages 45–73 years) and absent in other nine unaffected family members (Fig. 3e). All studied individuals in Fam-59 were homozygous for APOE ɛ3.
MARCH4 p.(Lys211Glu) variant is present in ExAC as a singleton in a non-Finnish European sample and in SISu as a singleton (identifier: rs756981946). In silico predictions gave the following results: Polyphen2 predicted the variant to be probably damaging (score 0.995), SIFT tolerated (score 0.29), MutationTaster damaging (score 1.000), MutationAssessor medium effect (score 2.22), CADD Phred-like scaled C-score was 19.62. These data demonstrate that the variant is extremely rare and suggest that it might alter the normal function of MARCH4.
The p.(Trp13Cys) variant is more common as it is reported in ExAC with a frequency of 0.001285 in Finnish samples. It has also been detected in other populations: European (3/56914), South Asian (8/13234), African (3/7972) and Latino (1/9762). SIFT predicted this variant to be tolerated (score 0.29), Polyphen2 benign (score 0.00), MutationTaster damaging (score 0.981), MutationAssessor neutral (score −0.55) and CADD Phred-like scaled C-score was 2.416. These predictions along with the fact that it was present in individuals over 70 years of age suggest that p.(Trp13Cys) might be a rare neutral variant.
Discussion
In contrast to EOAD, LOAD is rarely caused by segregating variants in families. The strongest identified risk factor is the APOE ɛ4 allele. In few cases, variants in APP, PSEN1 and PSEN2 have been reported in LOAD families [9]. GWAS studies have identified ~20 loci associated with predisposition to AD but finding variants that actually have a biological effect has proven difficult. In FTD, variants in C9orf72, MAPT and GRN account for up to 60% of familial cases while variants in other genes are rare [11].
In addition to the ALS/FTD entity, C9orf72 expansions have been linked to several other clinical manifestations including AD, Parkinson’s disease and Huntington’s disease phenocopies (reviewed in ref. [38]). We detected C9orf72 expansions in 7/60 (11.6%) families with either FTD or FTD/ALS but also in 3/60 (5%) families with clinical AD. In one family, Fam-31, neuropathological examinations disclosed moderate to severe AD tau-pathology, and no TDP-43-positive inclusions were seen. However, p62-positive inclusions were present in the cerebellum, consistent with the C9orf72 expansion. Several earlier reports have described C9orf72 expansions in either clinically diagnosed [39,40,41,42] or neuropathologically confirmed AD [43]. It is possible that the AD pathology is at least partly attributable to APOE as the one affected individual with C9orf72 expansion and AD-type neuropathology was homozygous for the APOE ε4 allele (Supplementary table 2). Previous work has shown that C9orf72 expansions are seen in ~30% of Finnish FTD patients [13] and in 48.1% of familial FTD [44]. Our results confirm this finding and suggest that C9orf72 expansions may manifest as clinical AD and some patients may also show concomitant AD pathology at the neuropathological examination. Previous studies on C9orf72 expansions in AD patients have suggested that the clinical or neuropathological classification as AD may have been incorrect, and this appeared to be the case in some of our families with clinical diagnosis of AD.
No variants in APP exons 16 and 17 or the coding regions of PSEN1 and PSEN2 were observed in the 10 AD families selected from our cohort. Whole-genome genotyping also showed no clearly causative CNVs. Both results are in agreement with previous studies. Only a few PSEN1 variants have been reported in Finnish AD families: two families carry the ‘Cotton-wool’ variant, Δ9Finn (c.869_955del) [45], p.(Met146Val) has been reported in a Swedish family of Finnish descent [46, 47] and p.(Pro264Leu) in one family [48]. Screening of APP, PSEN1 and PSEN2 in a cohort of 140 EOAD patients revealed no variants that might affect function [49]. In addition, duplication of APP was not detected in a cohort of 64 Finnish EOAD patients [50].
GRN sequencing and exome sequencing did not reveal any pathogenic variants in the two FTD families without C9orf72 expansions. In agreement with our results, previous work suggests that GRN variants are rare among Finnish FTD patients [51].
Exome sequencing revealed rare, potentially relevant variants in five families. Two variants were in genes previously linked to AD (CLU and PCDH11X) while three variants were in genes (UNC13C and MARCH4) that have not been directly linked to dementia but could be important in maintaining normal neuronal function.
In 2010, a large GWAS study indicated that PCDH11X was linked to LOAD in a combined American Caucasian cohort [52]. However, subsequent studies in different populations failed to confirm the findings of Carrasquillo et al. [53,54,55,56] Recently, Jiao et al. [57] reported a single-nucleotide polymorphism in PCDH11X to confer a risk to LOAD. Thus, the possible role of PCDH11X in AD susceptibility is still somewhat unclear. Our results show that the rare p.(Asp760Val) variant is present in all affected individuals of family Fam-15 but also in two asymptomatic individuals and in one subject with unclear status.
The role of CLU as an AD risk gene has been established in independent data sets [3, 4]. We noted co-segregation of a rare CLU variant and dementia in an AD family (Fam-56). Even though rare non-synonymous and small insertion/deletion variants have been reported to increase AD risk [58, 59], the p.(Thr203Ile) variant is predictably not deleterious, but at present, we cannot exclude its possible role in AD risk.
Two AD patients from family Fam-49 shared a 3 bp in-frame deletion in UNC13C. The UNC13C gene is highly expressed in brain. Experimental evidence from cat and mouse models have suggested that its mammalian homologue, Munc13-3, has a role in controlling critical-period neuronal plasticity in visual cortex [60, 61]. Gene expression studies in human AD and control brain samples showed increased UNC13C expression in hippocampal CA3 compared to CA1 in Alzheimer patients. This implicates that UNC13C might have a neuroprotective role in the brain [62]. The rare variant found in family 49 removes one amino acid residue but does not disturb the reading frame. Both affected patients were also heterozygous for the APOE ɛ4 allele, a likely risk factor in this family.
A rare segregating missense variant in MARCH4 was identified in the FTD family Fam-13. MARCH4 is a member of membrane-associated RING-CH family of ubiquitin E3 ligases. These ligases function in the last step of ubiquitination by recruiting the ubiquitin carrying E2 enzyme and transferring ubiquitin from E2 to the target protein [63]. MARCH4 is predominantly expressed in the adult human brain [64]. The ubiquitin-related protein degradation pathway has been implicated in many neurodegenerative diseases, including FTD. Recent work by Williams et al. [27] described variants in a component of the ubiquitin E3 ligase complex, CCNF, in a large ALS/FTD family and a few singleton patients. Although the MARCH4 variant segregated with FTD in our small family, we cannot exclude the possibility that we merely identified a rare neutral variant in a gene with function that could fit in the model of FTD pathogenesis.
C9orf72 repeat expansions are common among Finnish FTD patients and our results indicate that expansions may also be seen in patients with clinical and neuropathological diagnoses of AD. Our results suggest that unknown genetic factors are likely to be responsible for a proportion of familial dementia in the Finnish cohort, but definitely causal or risk variants in novel genes are yet to be identified. Exome sequencing is an efficient way to search for rare coding variants, but thus far only few segregating risk variants (eg, TREM2 p.(Arg47His) [65] and TTC3 p.(Ser1038Cys) [66]) have been described in LOAD families. Our results corroborate the view that even in large LOAD families with multiple affected individuals, the disease is likely caused by combination of multiple genetic and environmental risk factors. The APOE ε4 risk allele can be assumed to account for multiple affected individuals in several of the AD families in our study.
We detected rare segregating coding variants in UNC13C in an AD family and in MARCH4 in an FTD family. However, replication in larger familial and case–control data sets and functional assays would be needed to prove their causality. The limitation of our study is the relatively small number of patients. Thus, we could only aim to find highly penetrant pathogenic variants. In addition, exome sequencing does not enable the identification of non-coding variants that might affect splicing or gene expression and repeat expansions are not reliably detectable.
While our exome sequencing approach failed to identify any clearly causal variants in the 12 families, we believe that the rare variants found in our cohort will be of interest for other dementia researchers. Thus, we presented all the variants and genes of potential interest in the hope this may be useful for future studies and can facilitate analyses in other families and data sets.
References
McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9.
Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–74.
Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–9.
Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–93.
Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–40.
Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–35.
Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–41.
Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8.
Lee JH, Kahn A, Cheng R, et al. Disease-related mutations among Caribbean Hispanics with familial dementia. Mol Genet Genomic Med. 2014;2:430–7.
Vardarajan BN, Ghani M, Kahn A, et al. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol. 2015;78:487–98.
Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet. 2015;386:1672–82.
Rohrer JD, Guerreiro R, Vandrovcova J, et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology. 2009;73:1451–6.
Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68.
DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56.
Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9.
Cruts M, Gijselinck I, van der Zee J, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4.
Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–5.
Benajiba L, Le Ber I, Camuzat A, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–3.
Van Langenhove T, van der Zee J, Sleegers K, et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–71.
Huey ED, Ferrari R, Moreno JH, et al. FUS and TDP43 genetic variability in FTD and CBS. Neurobiol Aging. 2012;33:1016.e9–17.
Watts GD, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–81.
Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–8.
Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5.
Synofzik M, Maetzler W, Grehl T, et al. Screening in ALS and FTD patients reveals 3 novel UBQLN2 mutations outside the PXX domain and a pure FTD phenotype. Neurobiol Aging. 2012;33:2949.e13–7.
Gijselinck I, Van Mossevelde S, van der Zee J, et al. Loss of TBK1 is a frequent cause of frontotemporal dementia in a Belgian cohort. Neurology. 2015;85:2116–25.
Le Ber I, Camuzat A, Guerreiro R, et al. SQSTM1 mutations in French patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol. 2013;70:1403–10.
Williams KL, Topp S, Yang S, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016;7:11253.
Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15:857–68.
Myllykangas L, Polvikoski T, Sulkava R, et al. Genetic association of alpha2-macroglobulin with Alzheimer’s disease in a Finnish elderly population. Ann Neurol. 1999;46:382–90.
MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–92.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2.
Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Guerreiro RJ, Beck J, Gibbs JR, et al. Genetic variability in CLU and its association with Alzheimer’s disease. PLoS ONE. 2010;5:e9510.
Chi S, Jiang T, Tan L, Yu JT. Distinct neurological disorders with C9orf72 mutations: genetics, pathogenesis, and therapy. Neurosci Biobehav Rev. 2016;66:127–42.
Cacace R, Van Cauwenberghe C, Bettens K. et al. C9orf72 G4C2 repeat expansions in Alzheimer’s disease and mild cognitive impairment. Neurobiol Aging. 2013;34:1712.e1–7.
Saint-Aubert L, Sagot C, Wallon D, et al. A case of logopenic primary progressive aphasia with C9ORF72 expansion and cortical florbetapir binding. J Alzheimers Dis. 2014;42:413–20.
Harms M, Benitez BA, Cairns N, et al. C9orf72 hexanucleotide repeat expansions in clinical Alzheimer disease. JAMA Neurol. 2013;70:736–41.
Majounie E, Abramzon Y, Renton AE, et al. Repeat expansion in C9ORF72 in Alzheimer’s disease. N Engl J Med. 2012;366:283–4.
Kohli MA, John-Williams K, Rajbhandary R, et al. Repeat expansions in the C9ORF72 gene contribute to Alzheimer’s disease in Caucasians. Neurobiol Aging. 2013;34:1519.e5–12.
Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–30.
Prihar G, Verkkoniem A, Perez-Tur J, et al. Alzheimer disease PS-1 exon 9 deletion defined. Nat Med. 1999;5:1090.
Alzheimer’s Disease Collaborative Group. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat Genet. 1995;11:219–22.
Hiltunen M, Helisalmi S, Mannermaa A, et al. Identification of a novel 4.6-kb genomic deletion in presenilin-1 gene which results in exclusion of exon 9 in a Finnish early onset Alzheimer’s disease family: an Alu core sequence-stimulated recombination? Eur J Hum Genet. 2000;8:259–66.
Martikainen P, Pikkarainen M, Pöntynen K, et al. Brain pathology in three subjects from the same pedigree with presenilin-1 (PSEN1) P264L mutation. Neuropathol Appl Neurobiol. 2010;36:41–54.
Krüger J, Moilanen V, Majamaa K, Remes AM. Molecular genetic analysis of the APP, PSEN1, and PSEN2 genes in Finnish patients with early-onset Alzheimer disease and frontotemporal lobar degeneration. Alzheimer Dis Assoc Disord. 2012;26:272–6.
Blom ES, Viswanathan J, Kilander L, et al. Low prevalence of APP duplications in Swedish and Finnish patients with early-onset Alzheimer’s disease. Eur J Hum Genet. 2008;16:171–5.
Krüger J, Kaivorinne AL, Udd B, Majamaa K, Remes AM. Low prevalence of progranulin mutations in Finnish patients with frontotemporal lobar degeneration. Eur J Neurol. 2009;16:27–30.
Carrasquillo MM, Zou F, Pankratz VS, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009;41:192–8.
Beecham GW, Naj AC, Gilbert JR, Haines JL, Buxbaum JD, Pericak-Vance MA. PCDH11X variation is not associated with late-onset Alzheimer disease susceptibility. Psychiatr Genet. 2010;20:321–4.
Lescai F, Pirazzini C, D’Agostino G, et al. Failure to replicate an association of rs5984894 SNP in the PCDH11X gene in a collection of 1222 Alzheimer’s disease affected patients. J Alzheimers Dis. 2010;21:385–8.
Wu ZC, Yu JT, Wang ND, et al. Lack of association between PCDH11X genetic variation and late-onset Alzheimer’s disease in a Han Chinese population. Brain Res. 2010;1357:152–6.
Miar A, Alvarez V, Corao AI, et al. Lack of association between protocadherin 11-X/Y (PCDH11X and PCDH11Y) polymorphisms and late onset Alzheimer’s disease. Brain Res. 2011;1383:252–6.
Jiao B, Liu X, Zhou L, et al. Polygenic analysis of late-onset Alzheimer’s disease from mainland China. PLoS ONE. 2015;10:e0144898.
Bettens K, Brouwers N, Engelborghs S, et al. Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk. Mol Neurodegener. 2012;7:3.
Bettens K, Vermeulen S, Van Cauwenberghe C, et al. Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations. Mol Neurodegener. 2015;10:30.
Yang CB, Zheng YT, Li GY, Mower GD. Identification of Munc13-3 as a candidate gene for critical-period neuroplasticity in visual cortex. J Neurosci. 2002;22:8614–8.
Yang CB, Kiser PJ, Zheng YT, Varoqueaux F, Mower GD. Bidirectional regulation of Munc13-3 protein expression by age and dark rearing during the critical period in mouse visual cortex. Neuroscience. 2007;150:603–8.
Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med. 2013;5:48.
Chaugule VK, Walden H. Specificity and disease in the ubiquitin system. Biochem Soc Trans. 2016;44:212–27.
Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Früh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol. 2004;78:1109–20.
Korvatska O, Leverenz JB, Jayadev S, et al. R47H variant of TREM2 associated with Alzheimer disease in a large late-onset family: clinical, genetic, and neuropathological study. JAMA Neurol. 2015;72:920–7.
Kohli MA, Cukier HN, Hamilton-Nelson KL, et al. Segregation of a rare TTC3 variant in an extended family with late-onset Alzheimer disease. Neurol Genet. 2016;2:e41.
Acknowledgements
Professors M. Haltia and R. Sulkava are acknowledged for their valuable contribution in the collection of the families. This study was financially supported by the Päivikki and Sakari Sohlberg Foundation (L.M. and P.P.), the Academy of Finland, grant number 294817 (L.M.), the Pirkko and Veikko Mäkelä foundation (P.P.), and Finnish Cultural Foundation, Kymenlaakso Regional Fund (P.P.), Sigrid Juselius Foundation (P.J.T.) and Helsinki University Hospital (P.J.T. and L.M.). This work was supported in part by the Alzheimer’s Society and Alzheimer’s Research UK. J.B. and R.G. are supported by fellowships from the Alzheimer’s Society. This study is dedicated to the memory of study nurse and friend Raija Ahlfors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Pasanen, P., Myllykangas, L., Pöyhönen, M. et al. Genetics of dementia in a Finnish cohort. Eur J Hum Genet 26, 827–837 (2018). https://doi.org/10.1038/s41431-018-0117-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0117-3