Abstract
It is currently unclear if next-generation sequencing (NGS) technologies can be implemented in the diagnosis setting at an affordable cost. The aim of this study was to measure the total cost of performing NGS in clinical practice in France, in both germline and somatic cancer genetics.
The study was performed on 15 French representative cancer molecular genetics laboratories performing NGS panels’ tests. The production cost was estimated using a micro-costing method with resources consumed collected in situ in each laboratory from a healthcare provider perspective. In addition, we used a top–down methodology for specific post-sequencing steps including bioinformatics, technical validation, and biological validation. Additional non-specific costs were also included. Costs were detailed per step of the process (from the pre-analytical phase to delivery of results), and per cost driver (consumables, staff, equipment, maintenance, overheads). Sensitivity analyses were performed.
The mean total cost of NGS for targeted gene panels was estimated to 607€ (±207) in somatic genetics and 550€ (±140) in germline oncogenetic analysis. Consumables were the highest cost driver of the sequencing process. The sensitivity analysis showed that a 25% reduction of consumables resulted in a 15% decrease in total NGS cost in somatic genetics, and 13% in germline analysis. Additional costs accounted for 30–32% of the total NGS costs.
Beyond cost assessment considerations, the diffusion of NGS technologies will raise questions about their efficiency when compared to more targeted approaches, and their added value in a context of routine diagnosis.
Similar content being viewed by others
Introduction
Molecular diagnostics in cancer have dramatically changed in recent years [1]. For the past 30 years, Sanger sequencing technology has been the gold standard for the detection of germline genetic variants and has been accepted as a reference for somatic genetic variants for which other very sensitive methods have been developed (qPCR, pyrosequencing…) [2]. Next-generation sequencing (NGS) is a breakthrough replacing these conventional molecular diagnostic technologies, allowing higher throughput analyses, improved cost effectiveness and speed, and gained in both sensitivity and specificity [3, 4]. As cancer is a multiparametric molecular disease, the simultaneous screening of a large number of genes should be intuitively more effective than a gene-by-gene approach. While NGS technologies were originally developed in a research context, their transfer to clinical diagnostics has been rapidly performed with laboratory developed techniques [5, 6]. Therefore, it is difficult to correctly assess the gain in production toward the cost.
For more than 10 years, molecular diagnostic tests in France have been performed nationwide in a setting of a network of 28 platforms and 25 germline oncogenic laboratories, supported by the French National Cancer Institute (INCa) and the Ministry of Health. They are free of charge for patients, giving equal access to genomic-based cancer care throughout France. In 2015, 75,000 cancer patients benefited from predictive molecular tests with 121 tests being performed in France, including 15,000 NGS tests [7]. This network in place has eased the diffusion of the NGS testing.
Implementation of NGS in clinical laboratories is challenging. Beyond the question of quality of these tests compatible with its clinical use, these new techniques emerge in the context of increasing concerns about healthcare costs because these tests are aimed to guide the prescription of expensive targeted therapies or immunotherapies [8,9,10,11,12,13]. The cost of sequencing per base has decreased 5-fold in the last 10 years [14], and commercial producers are actually performing whole-genome sequencing for less than $1000 [15,16,17].
Whilst this has rendered NGS more affordable, the deployment of NGS on existing molecular platforms with a progressive shift toward targeted NGS for all patients still needs economic evaluation before its national diffusion.
Economic evaluation of genomic sequencing is however challenging [18, 19]. Most of the studies evaluating the economic impact of NGS [11, 20] only considered the equipment and consumable costs for library preparation and sequencing. Most of the time they did not include costing for ‘non-production’ activities such as quality assessment or the parallel development of tools for improving sequencing pipelines and bioinformatic analysis. The reagents costs are insufficient to really appreciate the real cost of this new approach. Very few studies have estimated a complete cost of NGS for routine clinical diagnosis using observational multicenter data [21].
The aim of this study was to measure the total cost of performing NGS of routine clinical samples (both somatic and germline cancer genetic analysis) from 15 representative cancer molecular genetic laboratories currently performing NGS in France.
Materials and methods
Laboratories included in this study
Our study included data from 15 French genetics laboratories performing somatic and germline clinical NGS tests. The laboratories were chosen because they performed cancer diagnostics nationwide and presented the full spectrum of clinical indications and technical solutions.
Costing methodology
The production cost for all samples included in one NGS run was determined using a micro-costing method [22]. Resources consumed were collected in situ for each laboratory, using a prospective direct observation using a standardized questionnaire.
Observations were investigated from March 2014 to November 2015.
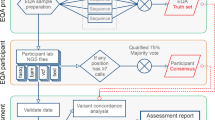
The production costs were calculated for the overall process, from the pre-analytical phase (sample collection and DNA extraction), enrichment/library preparation (including technical failures), sequencing, bioinformatics analysis, technical validation, and biological validation. Technical validation referred to the tests performed to confirm variants discovered by NGS. Biological validation is the clinical interpretation of variants, that is classifying the variants and reporting them in a way that is meaningful for the clinician.
The time horizon was the period from registration of the sample until the delivery of the results to the clinician. Costs were not discounted. Costs were calculated from the perspective of each NGS platform involved in the study (healthcare provider perspective).
In addition to micro-costing, we performed an activity-based costing using top–down approach for specific post-sequencing steps including bioinformatics, technical validation, and biological validation. Bioinformatics and its related pipeline development costs were assessed by two laboratories, and were considered as the minimum investment amount to develop and implement NGS routine tests. DNA extraction micro-costing was calculated from data collected from two laboratories.
We identified resources consumed for each step of the NGS process: personnel (labor time), consumables (i.e., reagents and other consumables), and equipment (usage and maintenance). Personnel costs were calculated using the time spent on each task by each category of staff. This time was valued using a mean gross wage of each category of personnel involved. We assumed, according to the French Labour Legislation, that the annual working times of personnel was 35 hours a week. Consumable costs were calculated based on real consumption and real unit purchases prices. Equipment costs were calculated based on their utilization time multiplied by their hourly costs. Hourly costs for equipment were calculated using the acquisition price without discount, on the basis of 2016 h (252 days, 8 h/day) of use per year and 6048 h for MiSeq (252 days, 24 h/day) with a 5-year linear amortization. Hourly costs for maintenance were calculated using the annual contract maintenance fee divided by 1820 h of use per year. If not available, a fixed price of 10% of the unit price without discount was applied.
We also measured the additional non-specific costs associated with routine clinical sample NGS:
-
Cost of personnel time required, but not specifically related to NGS activity (training, meeting to implement NGS…). This was assigned at 35% of total personnel cost.
-
Overhead costs (e.g., logistic expenses, water, energy, administration expenses…) were assigned at 27% of the total cost according to the French National Hospital Accounting Unit (ENCC for Etude Nationale des Coûts à Méthodologie Commune). It is a French hospital cost database which provides detailed cost information for hospital stays.
-
Other additional costs included the cost of routine accreditation and certification, cost of R&D (Research and Development) in clinical practice corresponding to constant improvement in our technical application, and other fees (waste treatment, sample shipping fees). This was assigned at 3% of the total gross cost, estimated from activity-based data from one laboratory.
Results are presented as a total cost per patient (cost per sample) for each laboratory, by separating somatic and germline applications, and are also detailed by steps. The NGS production cost will first be presented (without overhead) and then the total cost including overheads will be computed.
Costs are expressed in 2014 euros (€ VAT). All statistical analyses were performed using STATA software (version 12.1). Uncertainty was captured using deterministic sensitivity analyses.
Results
Laboratories’ characteristics
The micro-costing analysis was performed for seven somatic genetics (solid tumor and oncohematology) and eight germline oncogenetic laboratories. Detailed characteristics of the indications and technologic choices of the laboratories are presented in Tables 1 and 2.
For somatic laboratories, there were considerable differences between the laboratories with respect to the number of patients analyzed per run (ranging from 7 to 70), the number of genes sequenced, the choice of the sequencing platform, and the enrichment technique (homemade vs. commercially purchased solutions/reagents). Enrichment techniques based on PCR (ampliseq) and sequencing platforms with PGM were used by the majority of laboratories performing somatic analyses.
For germline laboratories, we observed less heterogeneity in the technical choices, with five out of eight laboratories using a Miseq platform. The choice of enrichment technique was quite variable with five different technologies used.
Total NGS cost
A total cost per patient was calculated for each of the 15 laboratories. This cost is presented in Table 1 and Table 2 for somatic and germline genetic analyses, respectively.
Somatic genetic analysis
An overview of the total and individual component costs of the seven laboratories analyzed are shown in Table 1. The total costs ranged from 376€ to 968€ per patient, according to the laboratory.
The enrichment step accounted for the largest part of the NGS production cost for most of the laboratories, and was mostly driven by consumables representing between 89% and 97% of the cost of the step. The laboratory with the lowest enrichment cost (104€) used “home-made” solutions, whose cost was between 8% and 65% less expensive than the commercial solutions (Ampliseq and Sentosa).
The sequencing step was less expensive than the enrichment step for most laboratories, ranging from 24€ to 330€. This cost was also mostly driven by consumable costs (from 59 to 97% of this step). The largest cost heterogeneity between laboratories was observed in this step, the cost per sample being lower for a Proton Sequencer when compared to PGM. Acquisition equipment costs however accounted for only a small percentage of the per-sample sequencing cost (from 4 to 21% of the step).
The cost of bioinformatic analyses ranged from 33€ to 63€ per sample. This cost was mostly driven by equipment and software costs, accounting for 51–97% of the step.
The cost of technical validation was quite different between laboratories, ranging from 0€ (no validation) to 81€ (Sanger sequencing, and/or other tests). Some laboratories performed Sanger sequencing to confirm the variants detected by NGS, while others did not.
The cost of biological validation, which ranged from 12€ to 41€, only consisted of labor costs. The variability between laboratories was driven by the time spent by the biologist to interpret the biological relevance before reporting to the clinician. In some laboratories, a cross-validation by two biologists was performed.
The additional costs consisted of those not directly linked to NGS activity, including overhead costs and other additional costs (quality, R&D, and other fees). They accounted for a large proportion of the total cost, representing around 30%.
Germline oncogenetic analysis
An overview of the total and individual component costs of the eight laboratories analyzing targeted germline sequences are summarized in Table 2. The costs ranged from 322€ to 727€.
The enrichment step accounted for the largest NGS production cost for most laboratories, and was mostly driven by consumables required to prepare samples (from 81 to 95% of the step).
The sequencing step was less costly than the enrichment step for most laboratories, ranging from 18€ to 114€. This cost was also mostly driven by consumables costs (from 19 to 87 % of this step).
The cost of bio-informatic analyses ranged from 31€ to 62€ per sample, and represents between 6% and 11% of the NGS production cost. This cost was mostly driven by equipment and software costs accounting for 50–100%.
The cost of technical validation was quite different between laboratories, ranging from 10€ to 68€ depending on the technical test performed to confirm the variant. Some laboratories performed Sanger sequencing to confirm the variants detected by NGS. Overall, the cost of this step was equally divided between consumables and labor costs.
The cost of biological validation which ranged from 26€ to 105€ was only composed of labor costs. The variability between labs was driven by the time spent by the biologist to interpret the biological relevance before reporting to the clinician. In some labs, a cross-validation by two biologists was performed.
The additional costs accounted for a large proportion of the total cost, representing around 32% of the total.
Mean cost per cost driver in somatic and germline cancer genetics
The total NGS cost was on average 607€ (±207) for somatic genetic analysis and 550€ (±140) for germline genetic analysis (Tables 1 and 2). Consumable costs represented 48% of the total cost in somatic genetics and 41% in germline oncogenetics (Table 3). Equipment and software costs were 8% and 9% for somatic and germline analyses, respectively. Labor costs were 14% and 18%, respectively, in somatic genetic and germline genetic analyses. Finally, additional costs were between 30% and 32% of the total.
Sensitivity analysis
The results of the sensitivity analyses are presented in Figs. 1 and 2 for both somatic and germline genetic analyses, respectively.
The sensitivity analysis showed that a 25% reduction of consumables unit prices resulted in a 15% decrease in total NGS cost in somatic genetics, and 13% in germline analysis. Conversely, purchase equipment prices had a little impact on the total NGS cost (<1% for both somatic and germline genetics). A decrease of 25% in the cost of equipment’s would reduce the total cost per patient by €4 (€607−€603) and €5 (€550−€ 545) in somatic and germline genetics, respectively. A reduction in wages by ±25% altered the average costs by about 7% and 5%, respectively, in somatic genetics, and 7% and 9%, respectively, in germline genetics.
Discussion
Biologists and clinicians are now in an era where innovative technologies such as NGS can provide rapid high-throughput interrogation of complex germline and somatic alterations from clinical cancer patient samples. Due to the increasing demand for such tests and to reduce the time required to report results [23, 24], it has become important to provide a clear cost estimation associated with genome sequencing as a component of the total clinical care costs.
Our prospective study estimated the cost of NGS sequencing for targeted gene panels at 607€ (±207) on an average in somatic genetics and 550€ (±140) on an average in germline oncogenetics, in 15 laboratory settings.
In both somatic or germline genetic analyses, the enrichment phase is the most costly stage (29% and 34% of the cost of the sequencing process, respectively), whereas the sequencing phase itself represents only 20% and 9% of the process costs, respectively (Tables 1 and 2). This confirms that the enrichment step of generating the genomic DNA librairies is traditionally the most time-consuming and costly, and constitutes the major cost of DNA sequencing [11, 25].
Consumables were the highest cost driver of the sequencing process: 48% of the total NGS cost in somatic genetics and 41% in germline genetics. The additional costs, which consisted overhead cost, cost of time not directly linked to NGS activity, quality cost, R&D cost, and other fees, represented the second highest cost driver with 30% and 32%. This figure is high, and mainly explained by the fact that all institutions concerned are tertiary care centers which have an estimated 13% added cost for research and teaching. Labor costs were the third cost driver with 14% and 18% of the total cost in somatic and germline genetics, respectively. Importantly, the labor cost component is projected to increase significantly in the future due to more time being spent analyzing and interpreting the data. Conversely, it can be realistically expected that both equipment and consumable costs would decline due to economies of scale and competition. The sensitivity analysis indicated that the mean NGS cost was most sensitive to a variation of unit consumables prices, with a 25% reduction of consumables resulting in a 15% decrease in the total NGS cost in somatic genetics, and 13% in germline analysis.
Several studies have addressed the economic analyses of targeted gene panels [11, 17, 21, 25,26,27]; however, it has been difficult to compare studies which do not include the same cost components.
The majority of available economic studies have concentrated on the costs of the enrichment and sequencing steps alone, neglecting the other stages of the process.
The main strength of our study is to assess a complete, transparent, and up-to-date cost per patient, from DNA extraction to delivered results. Our study has taken into account additional costs which are not directly linked to NGS sequencing such as overhead costs, cost of time spent not directly linked to NGS activity (meeting, training, administrative time), additional cost of quality (certification, accreditation…), and cost to develop methods for clinical practice. These costs are rarely integrated in published cost analyses, except in the study by Sabatini et al. [21]. Our study demonstrates that these additional costs represent a significant part of the total NGS cost (30% and 32% in somatic and germline genetics, respectively). This means that specific infrastructure requirements (dedicated installations, skilled staff, up-to-date equipment, efficient software,…) and a high learning curve are needed to achieve NGS production.
Our study is also one of the few that includes development costs for bioinformatics pipeline analysis. We have estimated the time required for staff working in bioinformatics to analyze data and develop bioinformatic tools (resources for data storage, mining, and processing). The cost of bioinformatics is not often included in economic studies. This is despite numerous authors [28, 29] arguing that it is no longer possible to ignore this cost, and predicting that by 2020 the main cost associated with NGS will be generated by data analysis rather than data generation [30]. Muir et al. outlined that efforts to better incorporate detailed and realistic accounting for downstream bioinformatics analysis is important to the development of accurate model of full economic evaluation of sequencing projects.
Our study also measured the cost of technical validation. These validations were conducted only if relevant genetic alterations were identified by NGS. These variants were verified using traditional Sanger technology, which is the current gold standard.
Another strength of our study is that it is an observational and multicentric study and therefore reflects NGS practices in clinical cancer genetic applications. The national network of 28 regional platforms dedicated to molecular genetics was autonomously implemented in France. This has resulted in a large variety of technical solutions and practices, as well as differing levels of technical maturity and expertise between laboratories that we accounted for. However, one challenge is the difficulty in estimating a cost per sample in the context of wide differences and the non-consensus between laboratories (technology and investment in laboratory development or commercial bioinformatics, technical protocols, development, validation, …).
This results in the main methodological limitation of this work. In particular, the heterogeneity encountered in technical validation is due to the lack of consensus in the absence of national and/or international recommendations at the time of our study.
In the future, costs for technical validation are expected to decrease. This cost is currently high as the technique is newly implemented and there will be a minimal amount required to cover specific NGS validation requirements to ensure a high level of validation and quality control measures.
Assessment of the bioinformatics validation steps in the various laboratories also demonstrated a strong heterogeneity: between turnkey industrial solutions, academic solutions, and mixed solutions. The valorization of this stage, in particular, highlighted the difficulties of separating clinical routine activities from those of research development, as well as various activities of bio-informaticians involved.
In order to reduce inter-laboratory technical heterogeneity and to standardize in very large data sets, a network for high-throughput genome sequencing with a national data analysis center to perform data analysis and ensure consistency across the network of sequencing sites is being developed in France. It is projected that this network will be able to handle 235,000 genomes a year by 2020. A first relevant step will probably be to develop national guidelines and standardizing gene panels at the national level.
Despite the difficulties encountered by various practices and the level of technical maturity of the laboratories, it's very interesting to evaluate a process at a very early stage of development. As highlighted by our study, innovation is often analyzed in the context of non-stabilization from the process itself, and argues for upstream economic assessment to provide decision-makers first-hand data to guide early decisions in a constrained economic and budgetary context [31]. Early-stage economic analyses are essential for Governments and allied stakeholders to support coverage and determine reimbursement levels [32, 33]. This is somewhat complex in the case of NGS technologies because the healthcare system should be able to cope with the fast evolution of these tests [34].
While the number of routine clinical biomarkers is currently quite limited, NGS technologies have enormous potential, with future NGS applications most likely relying on wider analysis such as whole-exome and whole-genome sequencing.
The panels assessed in our study are currently limited to hotspot mutations for somatic panel and validated genes for germline panel. The cost of the interpretation is today quite limited. The extension of the number of target should increase the NGS cost.
Beyond cost assessment considerations, the diffusion of these new sequencing technologies will raise questions about their efficiency when compared to more targeted approaches, and their added value in a context of routine diagnosis [35, 36]. This work is a first step to provide an input for future cost-effectiveness analyses.
Change history
15 June 2018
Since the publication of the article, it has been noted that there is an error in Table 2. Where 543€ is listed in the final column of the table, this should have been written as 550€.
References
Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470:198–203.
Tsiati AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12:425–32.
Martinez DA, Nelson MA. The next generation becomes the now generation. PLoS Genet. 2010;6:e1000906.
Sikkema-Raddatz B, Johansson LF, de Boer EN, et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–42.
Boyd SD. Diagnostic applications of high-throughput DNA sequencing. Annu Rev Pathol. 2013;8:381–410.
Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80.
French National Cancer Institute (INCa). Molecular Genetics centres activity 2015. http://lesdonnees.e-cancer.fr/Themes/Soins/Les-tests-de-genetique-somatique.
Fineberg HV. Shattuck Lecture. A successful and sustainable health system--how to get there from here. N Engl J Med. 2012;366:1020–1027.
Armstrong K. Can genomics bend the cost curve? JAMA. 2012;307:1031–32.
Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275–80.
Christensen KD, Dukhovny D, Siebert U, Green RC. Assessing the costs and cost-effectiveness of genomic sequencing. J Pers Med. 2015;5:470–86.
Douglas MP, Ladabaum U, Pletcher MJ, Marshall DA, Phillips KA. Economic evidence on identifying clinically actionable findings with whole genome sequencing: a scoping review. Genet Med. 2016;18:111–6.
Hartman M, Martin AB, Lassman D, Catlin A, National Health Expenditure Accounts Team. National health spending in 2013: growth slows, remains in step with the overall economy. Health Aff. 2015;34:150–60..
Feero WG, Wicklund C, Veenstra DL. The economics of genomic medicine: insights from the IOM Roundtable on Translating Genomic-Based Research for Health. JAMA. 2013;309:1235–6.
Mardis ER. Anticipating the $1,000 genome. Genome Biol. 2006;7:112.
Hayden EC. Technology: the $1,000 genome. Nature. 2014;507:294–5.
Nimwegen KJM, van, Soest RA, van, Veltman JA, et al. Is the $1000 genome as near as we think? A cost analysis of next-generation sequencing. Clin Chem. 2016;62:1458–64.
Buchanan J, Wordsworth S, Schuh A. Issues surrounding the health economic evaluation of genomic technologies. Pharmacogenomics. 2013;14:1833–47.
Fugel H-J, Nuijten M, Postma M, Redekop K. Economic evaluation in stratified medicine: methodological issues and challenges. Front Pharmacol. 2016;7:113.
Frank M, Prenzler A, Eils R, Graf von der Schulenburg J-M. Genome sequencing: a systematic review of health economic evidence. Health Econ Rev. 2013;3:29.
Sabatini LM, Mathews C, Ptak D, et al. Genomic sequencing procedure microcosting analysis and health economic cost-impact analysis: a report of the association for molecular pathology. J Mol Diagn. 2016;18:319–28.
Tan SS, Rutten FFH, van Ineveld BM, Redekop WK, Hakkaart-van Roijen L, Comparing methodologies for the cost estimation of hospital services. Eur J Health Econ. 2009;10:39–45.
Evans JP, Dale DC, Fomous C. Preparing for a consumer-driven genomic age. N Engl J Med. 2010;363:1099–103.
Dietel M, Jöhrens K, Laffert MV, et al. A 2015 update on predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance. Cancer Gene Ther. 2015;22:417–30.
Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). www.genome.gov/sequencingcostsdata.
van Amerongen RA, Retèl VP, Coupé VM, Nederlof PM, Vogel MJ, van Harten WH. Next-generation sequencing in NSCLC and melanoma patients: a cost and budget impact analysis. Ecancermedicalscience. 2016;10:684.
Plöthner M, Frank M, von der Schulenburg J-MG. Cost analysis of whole genome sequencing in German clinical practice. Eur J Health Econ. 2017;18:623–33.
Mardis E, Raison C. Next-generation sequencing, cancer and molecular diagnostics: an interview with Elaine Mardis. Expert Rev Mol Diagn. 2015;15:449–50.
Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207.
Muir P, Li S, Lou S, et al. The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol. 2016;17. https://doi.org/10.1186/s13059-016-0917-0.
Crawford JM, Bry L, Pfeifer J, et al. The business of genomic testing: a survey of early adopters. Genet Med. 2014;16:954–61.
Deverka PA, Dreyfus JC. Clinical integration of next generation sequencing: coverage and reimbursement challenges. J Law Med Ethics. 2014;42:22–41.
Van den Bulcke M. Tests de Panels de gènes par Next Generation Sequencing pour un traitement ciblé en oncologie et en hémato-oncologie – Synthèse - KCE_240Bs_NGS_traitement_cible_Synthese.pdf. https://kce.fgov.be/sites/default/files/page_documents/KCE_240Bs_NGS_traitement_cible_Synthese.pdf. Accessed 13 Apr 2017.
Evans BJ. The limits of FDA’s authority to regulate clinical research involving high-throughput DNA sequencing. Food Drug Law J. 2015;70:259–87.
Bennette CS, Gallego CJ, Burke W, Jarvik GP, Veenstra DL. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015;17:587–95.
Phillips KA, Ann Sakowski J, Trosman J, Douglas MP, Liang S-Y, Neumann P. The economic value of personalized medicine tests: what we know and what we need to know. Genet Med. 2014;16:251–7.
Acknowledgements
We are grateful to all the staff of the laboratories involved in this study that participated in data collection, and we thank especially the laboratories that allowed site visits. In somatic genetics, these laboratories were: Gustave Roussy: Biopathology integrated genetic platform Molecular Pathology; Angers University Hospital; Plateforme SNP, Transcriptom et Epigénomique; Comprehensive Cancer Center Curie, Paris; Unité de Parmacogénomique; Service de Génétique; Rennes University Hospital; Cancer Somatic Genetics Laboratory; Lille University Hospital; Cancer Molecular Genetic Centre; Lille University Hospital; Laboratory of Oncohaematology; Department of Biology-Pathology-Genetics; Comprehensive Cancer Center Bergonié, Bordeaux; Molecular Pathology Unit. In germline genetics, these laboratories were: Lille University Hospital; Digestive Molecular Oncogenetic Unit; Comprehensive Cancer Centre CRLCC François Baclesse, Caen; Cancer Biology and Genetics Laboratory; Comprehensive Cancer Center Curie, Paris; Unité Génétique Constitutionnelle; Service de Génétique; Oscar Lambret Cancer Centre, Lille; Human Molecular Oncology Unit; Gustave Roussy; Biopathology integrated Genetic platform; Oncogenetic Laboratory; Lille University Hospital; Endocrine Molecular Oncogenetic Unit; Rouen University Hospital; Molecular Genetics, Department of Genetics; Nantes University Hospital; Plateforme de Génétique moléculaire des cancers; Service de génétique médicale. We are grateful to Frederique Nowak and Etienne Lonchamp (biology, transfer and innovations department, National Cancer Institute, France), Huong Ly Hoang (Institut Curie, Paris, France), Maroua Mimouni (URC-Eco, Ile-de-France, Paris, France), Dominik Heinz, and Marie Warren Collon (Cancer Centre Léon Bérard, Lyon, France).
Funding
This study was supported by a grant from the French National Cancer Institute, dedicated to economic analyses of innovative techniques (reference number 2013-1-NGS-02). This research was funded by the National Cancer Institute in France (INCa) and the Canceropôle Ile de France. The views expressed in this work are those of the authors and not those of the funding bodies.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Marino, P., Touzani, R., Perrier, L. et al. Cost of cancer diagnosis using next-generation sequencing targeted gene panels in routine practice: a nationwide French study. Eur J Hum Genet 26, 314–323 (2018). https://doi.org/10.1038/s41431-017-0081-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-017-0081-3
This article is cited by
-
Optimizing Patient Pathways in Advanced Biliary Tract Cancers: Recent Advances and a French Perspective
Targeted Oncology (2023)
-
A feasible molecular diagnostic strategy for rare genetic disorders within resource-constrained environments
Journal of Community Genetics (2023)
-
Cost of exome analysis in patients with intellectual disability: a micro-costing study in a French setting
BMC Health Services Research (2023)
-
EGFR-TKI re-administration after osimertinib failure in T790M mutation loss cases with re-biopsy
Investigational New Drugs (2022)
-
Evaluation of an inflammation-based score for identification of appropriate patients for comprehensive genomic profiling
Discover Oncology (2022)