Abstract
This study aims to identify gene defects in pediatric cardiomyopathy and early-onset brain disease with oxidative phosphorylation (OXPHOS) deficiencies. We applied whole-exome sequencing in three patients with pediatric cardiomyopathy and early-onset brain disease with OXPHOS deficiencies. The brain pathology was studied by MRI analysis. In consanguineous patient 1, we identified a homozygous intronic variant (c.850-3A > G) in the QRSL1 gene, which was predicted to cause abnormal splicing. The variant segregated with the disease and affected the protein function, which was confirmed by complementation studies, restoring OXPHOS function only with wild-type QRSL1. Patient 2 was compound heterozygous for two novel affected and disease-causing variants (c.[253G > A];[938G > A]) in the MTO1 gene. In patient 3, we detected one unknown affected and disease-causing variants (c.2872C > T) and one known disease-causing variant (c.1774C > T) in the AARS2 gene. The c.1774C > T variant was present in the paternal copy of the AARS2 gene, the c.2872C > T in the maternal copy. All genes were involved in translation of mtDNA-encoded proteins. Defects in mtDNA-encoded protein translation lead to severe pediatric cardiomyopathy and brain disease with OXPHOS abnormalities. This suggests that the heart and brain are particularly sensitive to defects in mitochondrial protein synthesis during late embryonic or early postnatal development, probably due to the massive mitochondrial biogenesis occurring at that stage. If both the heart and brain are involved, the prognosis is poor with a likely fatal outcome at young age.
Similar content being viewed by others
Introduction
In this study, we searched for the genetic defect in pediatric patients with severe cardiomyopathy and early-onset brain disease with deficiencies in oxidative phosphorylation (OXPHOS). This is often lethal at a very young age. The affected hearts were mostly thick-walled and diagnosed as hypertrophic cardiomyopathy (HCM). The brain pathology was investigated with magnetic resonance imaging (MRI). Additionally, muscle, liver, and developmental abnormalities were observed, this is not uncommon for OXPHOS diseases as the OXPHOS systems consists of five enzyme complexes, which generate ATP, the prime source of cellular energy, and OXPHOS defects often affect multiple organs and more particularly those organs with a high energy requirement. Overall, OXPHOS diseases have a prevalence of about 1 in 85,000. Genetically, 13 of the structural proteins of the OXPHOS complex are encoded by the mitochondrial DNA (mtDNA), which has a genetic code different from the nuclear DNA and therefore also contains the necessary transfer RNA (tRNA) and ribosomal RNA (rRNA) genes to generate mtDNA-encoded proteins [1]. The vast majority of the OXPHOS proteins, but also the proteins involved in replication, transcription, and translation of the mtDNA, in assembly of the OXPHOS complexes, and in mitochondrial quality control are encoded by hundreds of nuclear genes. This implies that these disorders can have a dual genetic origin [2]. The extreme variability of the clinical manifestations and the high number of genes potentially precludes genotype–phenotype correlations and complete, unbiased approaches, like whole-exome sequencing (WES), have become the preferred and successful strategy to reveal the underlying genetic cause.
We performed WES in three patients from unrelated families, one consanguineous and two non-consanguineous, in which mtDNA defects were excluded as cause. We filtered the consanguineous family for homozygous variants affecting protein function in regions of homozygosity (ROH), whereas for the non-consanguineous families first a targeted analysis of 412 mitochondrial disease genes and functionally or structurally related genes was performed [3]. When negative, this was followed by an analysis of all genes captured and sequenced. For both approaches, an autosomal recessive disease model was used [4,5,6,7,8]. In each family, a severe defect in a different gene was identified causing the disease, but all three genes were involved in translation of mtDNA-encoded proteins.
Materials and methods
Patients with pediatric cardiomyopathy, early-onset brain disease and OXPHOS deficiency
Patient I (Family I)
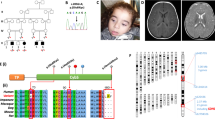
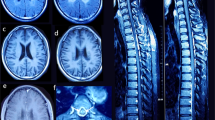
Patient 2.2, the female proband of a consanguineous Turkish couple, was prenatally diagnosed with HCM using fetal echocardiography (ultrasound) and postnatally confirmed by electrocardiography (ECG). From birth she developed encephalopathy, failure to thrive, and lactic acidosis. MRI of the brain at 5 months showed diffuse cerebral atrophy with T2 weighed imaging including hyper-intensive abnormalities in globus pallidus and putamen and also infratentorial in the dorsal brain stem, medulla oblongata, and mesencephalon, which is characteristic for Leigh syndrome (Fig. 1, the female patient 2.2). MRI spectroscopy of the white matter showed high lactic acid peak, and elevated biochemistry data for choline/creatine ratio, consistent with demyelination. She died at the age of 6 months. Her brother (2.1), with a comparable phenotype, died in utero and both were prenatally diagnosed with HCM using fetal echocardiography (ultrasound). Patient 2.2 was also prenatally diagnosed with a growth delay and a head lag. This head lag in infants is showing a developmental delay in head and neck muscle control. In conclusion, the pregnancy of 2.1 and 2.2 was not a normal development in contrast to 2.3 and 2.4.Their parents, a second sister (2.3), and the youngest brother (2.4) were healthy (Fig. 2a).
MRI of the brain data from patient I.2.2. aT1 weighted sagittal image: small corpus callosum, atrophy of the cerebellar vermis, abnormal intensity dorsal pons, and mesencephalic region (arrow). b Axial T2 weighted images showed subdural effusions, diffuse atrophy, frontal > occipital and hyperintensities in dorsal mesencephalon (arrow) in figure. c Hyperintensities in the dorsolateral part of the putamen (arrow). All three figures showed Leigh syndrome
Pedigree of three families with pediatric cardiomyopathy and Leigh (Family A) or Leigh-like syndrome (Family B and C) with multiple OXPHOS deficiencies. a Patient I.2.1 and I.2.2 from a consanguineous marriage carried a homozygous splicing defect in QRSL1 gene (NC_000006.11(NM_018292.4)). b Patient II.2.1 showed two novel variants in the MTO1 gene (NM_001123226.1). c Patient III.2.1, III.2.2, and III.2.3 showed two variants in the AARS2 gene (NM_020745.3)
The relative activities of OXPHOS enzyme complexes in muscle tissue biopsy of patient 2.2 were normalized to citrate synthase (CS); CI = 41%, CII = 68%, CIII = 70%, CIV = 71%, CV = 40%. Skin fibroblast (2.2) OXPHOS data showed CI = 43%, CII = 81%, CIII = 86%, CIV = 54%, CV = 64%. This indicates a complex I and V deficiency in muscle and a complex I, IV, and V deficiency in fibroblasts.
Patient 2 (Family II)
Patient 2.1 was diagnosed with a severe pediatric, HCM by ECG and died at the age of 1.5 year. He was the only child of non-consanguineous Dutch healthy parents (Fig. 2b) during a normal pregnancy. He showed diffuse cerebral atrophy with intraventricular septa on the MRI, characteristic for brain disease, and high levels of alanine and lactic acid, suggesting a mitochondrial disorder. The relative activities of OXPHOS enzyme complexes in muscle biopsy material of patient 2.1 were normalized to CS; CI = 11%, CII = 128%, CIII = 25%, CIV = 11%, CV = 61%, showing a severe deficiency of complex I, III, and IV. Skin fibroblast data showed a decreased activity of CI = 50%, unfortunately CII, CIII, CIV, and CV were not determined.
Patient 3 (Family III)
Patient 2.1 was born dysmature and was diagnosed at birth with severe HCM by ECG, and encephalomyopathy, persistent hypotonia, lung hypoplasia, and also high levels of lactic acid. Two other male siblings showed a similar phenotype and all three affected siblings died within 2 weeks after birth (2.2 within 1 day, 2.1 within 4 days, and 2.3 within 16 days) (Fig. 2c). The Dutch healthy parents were unrelated. Patient 2.1 and 2.2 patients were described as a spontaneous twin pregnancy and no complications before planned cesarean section. Nevertheless, after birth patient 2.2 showed immediately lung hypoplasia and died after 2 h post-mortem. No further information of patient 2.3 was available. Muscle biopsy (2.1) taken directly after birth showed many Cytochrome c oxidase (COX)-negative fibers with no other abnormalities. Postmortem brain pathology (2.1) confirmed the postmortem MRI and showed degeneration of the fasciculus gracilis and cuneatus with some vacuolization of the substantia nigra, characteristic for Leigh-like syndrome. The HCM was also confirmed during the postmortem heart pathology (2.1). The heart tissue was observed by transmission electron microscopy (TEM) and showed abnormal mitochondria, which were less in amount and large in size (data not shown). The relative activities of OXPHOS enzyme complexes in muscle tissue biopsy of patient 2.1 were normalized to CS; CI = 27%, CII = 120%, CIII = 11%, CIV = 6%, CV = 48%. Skin fibroblast data showed activities of CI = 45%, CII = 67%, CIII = 119%, CIV = 97%, CV = 107%. In muscle, all complexes showed a decreased activity, except complex II, whereas in fibroblasts only complex I was decreased.
Homozygosity mapping
Homozygosity mapping was performed using the Affymetrix GeneChip Human Mapping 250K SNP array (Affymetrix, Santa Clara, USA) using peripheral blood DNA of consanguineous parents and siblings (Family I). The DNA was labeled and processed according to the manufacturer’s instructions. Genotypes were generated by the Affymetrix Genotyping Analysis Software (GTYPE). Homozygous regions were determined by the online tool Homozygosity Mapper (http://www.homozygositymapper.org/).
WES and analysis
Genomic DNA was isolated from peripheral blood leukocytes using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). The quality of the genomic DNA was determined on a NanoDrop (Wilmington, USA) and by Agarose gel electrophoresis and finally quantified with PicoGreen on a Qubit fluorometer (Invitrogen, Bleiswijk, The Netherlands). A total of 4 µg starting material was used, which was further purified with a DNA Clean & Concentrator TM (ZymoResearch, BaseClear Diagnostics, Leiden, The Netherlands). The genomic DNA was fragmented to 150–200-bp lengths by shearing on a Covaris S2 (Covaris Inc., Massachusetts, USA). The exome was captured with the SureSelect Human All Exon V4 plus UTR’s kit (Agilent Technologies, Amstelveen, The Netherlands) for Paired-End Illumina Sequencing platforms (Illumina, Eindhoven, The Netherlands) using the standard SureSelect protocol (version 1.3). Quality was controlled on a Bioanalyzer 2100 (Agilent). Basecalling was performed using Casava 1.8 (Illumina). Reads were aligned to the human reference genome (UCSC hg19) using Burrows-Wheeler-Alignment tool (BWA) version 0.5.9. Single base variants were identified using Sequence-Alignment-MAP tools (SAM) version 0.1.16 and annotated according an in-house annotation Python/R script that matched variants to the RefGene, GenCode v19, and dbSNP144 hg19 tracks from the UCSC genome browser. Variants between the proband and the reference sequence were annotated and listed in an Excel file for further analyses. Two filtering strategies were applied. In the consanguineous patient I.2.2, homozygous variants from homozygosity regions were analyzed. In the non-consanguineous patients II.2.1 and III.2.1, first a diagnostic panel of 412 mitochondrial genes was analyzed for compound heterozygous variants, and if negative all captured genes would be analyzed.
All variants were filtered for a predicted functional effect (nonsense, missense, INDELs, and splice site variants) and frequency (<1% or absent in sequence databases (dbSNP144, Exome Variant Server, Exome Sequencing Project, Exome Aggregation Consortium (ExAC), and in-house patient database). Possible splice site variants were analyzed with Maximum Entropy Modeling of Short Sequence Motifs (MaxEnt), Neural Networks Berkeley Drosophila Genome Project (NNSPLICE), and Human Splicing Finder (HSF). Missense variants were classified, using SIFT (http://sift.jcvi.org/) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/). Additionally, in the consanguineous patient I.2.2, novel genes carrying variants were prioritized for their likelihood of being involved in mitochondrial disease using Weighted Gene Expression Tool and database (WeGET; http://weget.cmbi.umcn.nl). Shortly, remaining variants were analyzed for their co-expression with genes encoding mitochondrial proteins (mitochondrial system) in mammalian tissues. A weighted average of co-expression with mitochondrial system was calculated for all genes containing rare variants affecting protein function. A higher contribution was assigned to data sets that showed congruent expression (concomitant up and downregulation) of the mitochondrial system itself. The variants affecting the protein function could be the cause of the disease in the patient were submitted to the gene variant database. Links were described as: (1) www.lovd.nl/QRSL1, (QRSL1; #00100428: patient I.2.2), (2) www.lovd.nl/MTO1, (MTO1; #00100427: patient II.2.1), and (3) www.lovd.nl/AARS2, (AARS2; #00100426: patient III.2.1). Relevant variants were sequenced with gene-specific primers with an M13 tail and the BigDye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, Bleiswijk, The Netherlands) on an ABI 3730 DNA Analyzer. Data were processed using Sequence Scanner v1.0 from Applied Biosystems.
cDNA analysis and complementation assay of QRSL1
All primers used are listed in Table 1. RNA was isolated from patient and neonatal dermal human fibroblast cells (NDHF control cells) using Total RNA Isolation Kit according to the manufacturer’s manual (Roche Nederland B.V., Almere, The Netherlands). Copy DNA (cDNA) synthesis was performed by the qScript cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD, USA) using 1 µg of RNA input. Standard PCR steps: (1) 5 min 94 °C, (2) 1 min 94 °C, (3) 1 min 58 °C, (4) 1.5 min 72 °C, (5) 7 min 72 °C, repeat steps 2–4 34 cycles and cooling down to 4 °C. QPCR mRNA expression protocol were designed to generate a cDNA fragment across the site of the QRSL1 variant. QRSL1 mRNA expression was measured by the 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and was based on a primer pairs at exon 7 (209-bp amplicon), intron 7–exon 8 (169-bp amplicon, splice position), exon 8 (202-bp amplicon) and exon 9 (162-bp amplicon) of the QRSL1 transcript (NM_018292.4). QRSL1 expression was corrected for the TBP housekeeping gene (TATA box-binding protein, NM_003194.4, primer set in exon 5, 89-bp amplicon). Sensimix Sybr Hi-Rox (Bioline, Taunton, MA, USA) was used for cDNA amplification under the following conditions: 2 min 50 °C, 2 min 95 °C, followed by 40 cycles of 15 s 95 °C (denaturation) and 1 min 60 °C (annealing and extension). Nonspecific primer annealing was excluded by analysis of a primer dissociation curve, which was obtained by the following PCR steps: 15 s 95 °C, 15 s 60 °C, 15 s 60 °C, cooling down to 4 °C. For patient 1, QRSL1 mRNA expression was calculated using the standard 2-ΔΔ Ct method toward a TBP housekeeping gene, and compared with healthy control NDHF cells. The mutant spliced cDNA (NC_000006.11(NM_018292.4):c.850-3A > G) resulted in a percentage wild-type rest level of the normal spliced cDNA in exon 8. Mutant and wild-type PCR fragments were cloned using the pGEM T Easy cloning system (Promega, Madison, USA). Wild-type QRSL1 was expressed using QRSL1 cDNA (NM_018292.4) pcDNA3.1_DYK clone (Genscript, Piscataway, USA). All wild-type and mutant (patient 1 cDNA) QRSL1 constructs were also cloned in a pUltra-Chili Lentiviral (Addgene, Cambridge, USA) with special restriction enzymes sites primers (with an overhang generating restriction sites for BamHI (GGATCC) and XbaI (TCTAGA)).
Metabolic complementation experiments
Human fibroblasts were grown in media containing either glucose or galactose as the main carbohydrate source and the acidification rate or the oxygen consumption rate was determined with the Seahorse device (Seahorse Biosciences, Massachusetts, USA), according to the protocol of the manufacturer. Fibroblasts of patient I.2.2 and a control were transduced with wild-type and mutant (without exon 8) cDNA, cloned in a QRSL1-pUltra-Chilli Lentiviral vector [9]. The results for mitochondrial respiration are normalized to control fibroblasts, transduced with wild-type cDNA.
Results
Patient 1
For patient I.2.2, a total of 41,338 variants were detected in the WES data, of which 366 were homozygous variants mapping in the ROH. Variants that affected protein function were determined by in silico protein prediction tools, resulting in 24 of low frequency, damaging candidate variants (Supplementary Table 4). The genes involved were prioritized for their likelihood of causing mitochondrial disease using WeGET. Based on this prioritization list, we started our analysis with the QRSL1 gene, located in the ROH of chromosome 6, which carried a homozygous intronic variant A > G (NC_000006.11(NM_018292.4):c.850-3A > G (p.?), NP_060762.3), (Fig. 2a). The QRSL1 variant possibly leads to abnormal splicing, based on three splicing predictions (MaxEnt: –100.0%, NNSPLICE: –98.6% and HSF: –1.1%), which showed that the normal splice acceptor site was slightly reduced and an acceptor site 3-bp downstream could be used. Functional splicing validation was required and segregation analysis showed that both affected siblings (I.2.1 and I.2.2) were homozygous for the variant. The parents (I.1.1 and I.1.2) were heterozygous, as was the unaffected brother (I.2.4). The unaffected sister (I.2.3) did not carry the variant (Fig. 2a).
PCR amplification of the QRSL1 cDNA using primers spanning the site of the variant, followed by sequence analysis revealed three different PCR fragments of 439, 330 and 243 bp for the mutant, whereas the wild-type control cDNA showed only one band of 439 bp (Figs. 3a, b). Additional denaturing 10% UREA PAGE gel confirmed the presence of three different fragments, excluding the possibility of one band being a hetero-duplex (data not shown). Sequence analysis showed that in the PCR fragment of 243-bp exon 8 was completely absent (NM_018292.4:r.850_1042del), leading to a frameshift and premature stop codon at exon 9. In the PCR fragment of 330 bp, exon 8 was partly retained (37 amino acids of a total of 64 (NM_018292.4:r.850_931del), therefore missing 27 amino acids) and an exonic splice acceptor was used at position NM_018292.4:r.933 (c.933A). This also caused a frameshift and premature stop codon at position NM_018292.4:r.887* (c.887C). The 439-bp fragment was the normally spliced cDNA fragment. Quantitative PCR analysis showed that wild-type cDNA was about 40% of the amount observed in control fibroblast (Fig. 3c).
Analysis of splice defects in QRSL1 cDNA in patient I.2.2. a QRLS1 cDNA of patient I.2.2 with variant A > G (NC_000006.11(NM_018292.4):c.850-3A > G) showed three cDNA fragments. b Sequence analysis confirmed three different splicing events of intron 7 and 8, resulting in fragments of 439 bp (wild-type), 330 bp and 243 bp. b Percentage wild-type cDNA in patient I.2.2 compared with controls. The wild-type of patient I.2.2 cDNA (c.850-3A > G) resulted in a wild-type significant rest (*) level of 40% of the normal spliced cDNA in exon 8 (p < 0.05 by student T-test), three splice events for QRSL1 exon 8 in c
Metabolic complementation measurements in fibroblasts of patient I.2.2 showed defects in glycolysis and OXPHOS. For a complementation test, wild-type and mutant QRSL1 cDNAs was cloned in a pUltra-Chili Vector and subsequently used to transduce the fibroblasts of patient I.2.2 NDHF cells were negative controls. Patient cells transduced with wild-type cDNA QRSL1 showed a 61% OXPHOS activity (42% increase compared with non-transduced patient cells). The control cells showed a 51% OXPHOS activity when transduced with variant 1 (exon 8 was completely absent) cDNA QRSL1 (Fig. 4). Polybrene was added to enhance the expression after transduction, but in addition to that a minor cytotoxic effect was observed in all cells for all conditions.
Complementation of QRSL1 defects in fibroblasts with two different vectors. Transduction with QRSL1 cDNA without exon 8 showed a significant decrease of 49% ATP production in control cells. A significant increase of 42% ATP production in patient (I.2.2) and patient (I.2.2) plus transduced wild-type cDNA QRSL1 was observed. The standard deviation was measured at four independent time points after transduction (wild-type or mutant) and non-transduced (Control or Patient I.2.2). The results for mitochondrial respiration are normalized to control fibroblasts, transduced with wild-type cDNA. The * symbol indicates that the difference is statistically significant (student T-test).
Patient 2
In patient II.2.1, we detected 4540 variants in the 412 mitochondrial genes, of which the MTO1 gene, containing two novel variants (NM_001123226.1, NP_001116698.1), Alanyl-tRNA Synthase 10 (Supplementary Table 5), was the most likely candidate based on function and genotype–phenotype correlation [10, 11]. The patient carried a G > A substitution (NM_001123226.1:c.253G > A) in exon 2, changing p.(G85R), and a G > A substitution (NM_001123226.1:c.938G > A) in exon 5, changing p.(R313Q), both of which were damaging by in silico predictions of Polyphen (probably damaging, score 0.95) and Sorting Intolerant From Tolerant (SIFT-deleterious, score 0.001). The first variant was not present in the in-house and ExAC database, whereas the second variant (rs number 371179032) had a very low allele frequency (1/1211348, AF = 8.241E-6) in the ExAC database. Both variants were located in functional protein domains of MTO1, which were highly conserved, based on protein BioGraph annotated predictions. Each parent (II.1.1 and II.1.2) was heterozygous for one of the variants (Fig. 2b).
Patient III
In patient III.2.1, we detected 4917 variants in the 412 mitochondrial genes, of which we identified two unknown variants and one known variant in the AARS2 (NM_020745.3, NP_065796.1, Alanyl-tRNA synthetase) gene (Supplementary Table 6). Based on the HCM and Leigh-like phenotype, the AARS2 (Alanyl-tRNA synthetase) gene was the best candidate gene from the list (Supplementary Table 6). The first variant was a G > A substitution (NM_020745.3:c.1774C > T) in exon 13 (rs138119149 with a frequency of 0.001), causing a substitution of p.(R592W) with a damaging score based on Polyphen (probably damaging score 0.97) and SIFT (deleterious score 0.002). The first variant was listed in ExAC: rs138119149 (33/121110, AF = 0.0002725). The second variant was a G > A nonsense variant (NM_020745.3:c.2872C > T) in exon 22 had no rs number and was not present in the in-house or ExAC databases, p.(R958*). The unaffected father was heterozygous for the G > A (c.1774C > T) substitution in exon 13 and the unaffected mother was heterozygous for the nonsense variant c.2872C > T in exon 22 (Fig. 2c).
Discussion
Cardiomyopathy is a disease of the heart muscle, interfering with efficient contraction and often leading to heart failure. According to the definition of the American Heart Association (AHA), cardiomyopathies can be either primary limited to the heart or can be a secondary result of other illnesses [12]. In three families with pediatric cardiomyopathy, brain disease, and multiple OXPHOS deficiencies, we identified the causative variant by WES. Given the genetic heterogeneity of OXPHOS disorders with >1400 nuclear genes potentially involved, WES is the best suitable unbiased approach for finding the underlying genetic cause [13]. We started with an autosomal recessive disease model and in case of consanguinity as in patient I.2.2, we focused on homozygosity regions.
The second patient from a consanguineous marriage carried a homozygous splicing defect (NC_000006.11(NM_018292.4):c.850-3A > G, NP_060762.3) in the QRSL1 gene, leading to two mRNAs with a premature stop codon and a strong reduction of the normal mRNA. A complementation assay confirmed a causative role in the complex I deficiency and pathology. Therefore, we did not proceed with other candidate genes on the list. During the course of our studies, an article was published, reporting the first disease-causing variants in the QRSL1 gene. A homozygous variant (NM_018292.4:c.398G > T (p.(G133V)) was presented in a consanguineous family with two affected siblings (Patient Y and Z in Table 2) including HCM [14]. Also, a compound heterozygosity for the variants (NM_018292.4:c.350G > A (p.(G117E) and NM_018292.4:c.398G > T (p.(G133V)) was reported in another non-HCM patient (Patient AA in Table 2) with interstitial pneumonia and mitochondrial respiration deficiencies [14]. These data confirmed the involvement of QRSL1 in pediatric cardiomyopathy with OXPHOS deficiency. The clinical manifestations vary, and based on the limited data, a strong reduction in QRSL1 seems more severe than the amino-acid changes reported, although data on the brain phenotype are lacking (Tables 2 and 3).
QRSL1 (or GatA) is part of a heterotrimeric enzyme complex GatCAB; GatA, GatB (PET112L), and GatC (GATC). GatA (QRSL1) plays a role as a mitochondrial amidotransferase, recognizing Glu-tRNA (Gln) and converts Glu to Gln. An accumulation of Glu-tRNA previously showed defects of mitochondrial mammalian translation processes and respiration [15]. The QRSL1 defect can explain the observed defects in glycolysis and OXPHOS. This is supported by earlier data on the role of mitochondrial translation and glutaminyl-mt tRNAGln in mouse cells, which demonstrated that mgatA produced a strong mitochondrial translational aminoacylation defect [16]. In Drosophila, null mutants of GatA gene, the homolog of QRSL1, caused larvae to grow slowly and die before pupariation [17].
As one (I.2.1) of the siblings of family 1 died in utero, this could reflect a similar developmental defect. Both studies support the essential role for glutaminyl-mt tRNAGln in mitochondrial protein synthesis and OXPHOS.
In patient II.2.1, we identified two novel variants in the MTO1 gene, which is involved in mitochondrial tRNA modification. MTO1 catalyzes the 5-carboxymethylaminomethylation of the uridine base in mitochondrial tRNA-Gln, tRNA-Glu, and tRNA-Lys, usually together with MTO2 (TRMU). A disease-causing role of MTO1 in mitochondrial patients with HCM (Tables 2 and 3) has been described before [10, 11]. The two variants (NM_012123.3:c.1282G > A (p.(Ala428Thr) and NM_012123.3:c.1858dup (p.(Ala620Lysfs*8)) in siblings patients P and Q showed early-onset lethal HCM, but normal brain anatomy (Table 2). An unrelated third consanguineous patient R with NM_012123.3:c.1282G > A (p.(Ala428Thr) showed HCM without lethality, and also normal brain development. In two consanguineous families (lethal male siblings T/U and non-lethal female siblings V/W), a homozygous variant (NM_012123.3:c.1232C > T (p.(Thr411Ile)) was reported causing early-onset HCM including abnormal brain morphology by MRI. Our two novel variants in MTO1 gene in patient II.2.1 (Table 2, patient X) displayed a similar early-onset HCM, a diffuse cerebral atrophy with an intraventricular septa, and multiple OXPHOS deficiencies, and lethality at 19 months.
In patients III.2.1, III.2.2, and III.2.3, we identified two variants in the AARS2 gene: NM_020745.3:c.1774C > T (p.(R592W), and NM_020745.3:c.2872C > T (p.(R958*). AARS2 consists of three functional domains; the N-terminal catalytic domain, the editing domain and the C-terminal (C-Ala domain). The AARS2 gene belongs to the class-II aminoacyl-tRNA synthase family and the protein is functional as a dimer. The first variant (c.1774C > T) on the paternal allele are in the editing domain of the AARS2 gene. The editing domain removes incorrectly charged amino acids and the first (c.1774C > T) variant is previously reported as a disease-causing variant associated with HCM and a poor prognosis [18, 19]. The second novel stop (c.2872C > T) variant on the maternal allele is located in the c-terminus (C-Ala domain) for the AARS2 gene and most likely leads to nonsense-mediated decay of the transcript. The presence of a reduced amount of the protein and the presence of two detrimental amino-acid substitutions in the other allele could, especially in dimers like AARS2, explain the more severe disease manifestation, of pediatric cardiomyopathy, Leigh-like syndrome, and multiple OXPHOS deficiencies. One of the patients in literature (patient D in Table 2) was compound heterozygous for the same missense mutation in combination with a frameshift mutation (NM_020745.3:c.[647dup];[1774C > T]), (p.[(C218Lfs*6)];[(R592W)]). The frameshift variant (p.(C218Lfs*6)) probably leads to nonsense-mediated decay of the transcript. Both variants resulted in cardiomyopathy and lethality after 3 months of birth [19, 20].
Different variants affecting protein function have been reported for the AARS2 gene resulting in HCM, leukodystrophy, and ovarian failure [20, 21] and a genotype–phenotype correlation is needed for novel variants in genes. Structural modeling of tissue-specific mitochondrial alanyl-tRNA synthetase (AARS2) defects was previously reported to predict different effects on aminoacylation activity level for the protein [20]. The study showed a clear correlation between the amino-acid change (p.(R592W)) and the severity of the predicted aminoacylation activity of AARS2 linked to cardiomyopathy. In case of a significant residual amount of mischarged amino acids, the predicted activity of aminoacylation resulted in a brain disease phenotype. Strikingly, none of the missense variants in leukodystrophy patients were reported in the editing domain in contrast to the cardiomyopathy patients [20]. Nevertheless, several missense variants were described outside the editing domain (N-terminal catalytic and C-terminal (C-Ala domain) and caused a cardiomyopathy phenotype [18,19,20]. Further insights in protein modeling and in vivo studies (e.g., mice or zebrafish) can further elucidate the role of translation defects during mitochondrial protein synthesis.
In all three patients, pediatric cardiomyopathy, early-onset brain disease, and multiple OXPHOS deficiencies were present. Our data, together with other reports, indicates that the heart is sensitive to severe defects in mitochondrial translation during prenatal development (Table 3). The heart muscle consists for 30% of mitochondria, which have a critical role in the structure alignment of heart muscle fibers and, in supplying the heart with sufficient energy. Adequate mitochondrial translation is essential for both processes, especially during development when the heart differentiates from early embryonic cells by massive proliferation of the mitochondria. It is therefore likely that the disturbed translation causes structural problems (HCM) in combination with OXPHOS defects in the prenatal and neonatal heart. Evidence supporting this hypothesis can be derived from other cardiomyopathy disease-causing variants in mitochondrial translation, like the tRNAGly NC_012920.1:m.9997T > C, tRNAHis NC_012920.1:m.12192G > A, tRNALys NC_012920.1:m.8344A > G, tRNALeu NC_012920.1:m.3243A > G and NC_012920.1:m.3302A > G, tRNAIle NC_012920.1:m.4317A > G, tRNATrp NC_012920.1:m.5545C > T, and tRNAVal NC_012920.1:m.1644G > A. Furthermore, other defects in nuclear genes involved in mitochondrial protein synthesis are reported to cause pediatric cardiomyopathy, for example, mitochondrial ribosomal subunits (MRPL3, MRPL44), tRNA-modifiers (GTPBP3), and RNA processing enzyme (ELAC2) [20, 22]. Cardiomyopathy can be the sole manifestation, but it can also be part of a broader syndromic spectrum. However, not all defects in genes involved in mitochondrial translation lead to HCM, as apart from the AARS2 gene none of the tRNA synthetases genes affect the heart, but rather the brain, liver, and peripheral nervous system (Table 3). The number of variants involved in translation of mitochondrial proteins, is yet too low to define whether this is gene specific or variant specific, except for the AARS2 gene where a genotype–phenotype correlation has been established.
In conclusion, our study showed disease-causing variants in the nuclear aminoacyl-tRNA synthetase genes (AARS2, MTO1, QRSL1), causing pediatric cardiomyopathy, brain, and multiple OXPHOS deficiencies (Fig. 5). As the expression can be variable, it would be worthwhile to investigate the brain, even if the patient only presents with cardiomyopathy, as this is important for prognosis. Our data are in line with a key role of mitochondria in cardiovascular genetics and disease [23].
References
Gorman GS, Chinnery PF, DiMauro S, et al. Mitochondrial diseases. Nat Rev. Primer 2. Article number:16080. https://doi.org/10.1038/nrdp.2016.80.
Konovalova S, Tyynismaa H, Mitochondrial aminoacyl-tRNA synthetase in human disease. Mol Genet Metab. 2013;108:206–11. https://doi.org/10.1016/j.ymgme.2013.01.010.
Link or ref mitochondrial gene panel (412 genes); https://www.labmaastricht.nl/klinische-genetica
Gohil VM, Sheth SA, Nilsson R, et al. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat Biotechnol. 2010;28:249–55. https://doi.org/10.1038/nbt.1606.
Gerards M, Kamps R, van Oevelen J, et al. Exome sequencing reveals a novel Moroccan founder mutation in SLC19A3 as a new cause of early-childhood fatal Leigh syndrome. Brain. 2013;136(Pt 3):882–90. https://doi.org/10.1093/brain/awt013.
1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A map of human genome variation from population scale sequencing. Nature. 2015;526:68–74. https://doi.org/10.1038/nature15393.
Seelow D, Schuelke M, Hildebrandt F, Nurnberg P. HomozygosityMapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–9. https://doi.org/10.1093/nar/gkp369.
Ohtake A, Murayama K, Mori M, et al. Diagnosis and molecular basis of mitochondrial respiratory chain disorders: exome sequencing for disease gene identification. Biochim Biophys Acta. 2014;1840:1355–9. https://doi.org/10.1016/j.bbagen.2014.01.025.
Lou E, Fujisawa S, Morozov A, et al. Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS ONE. 2012;7:e33093. https://doi.org/10.1371/journal.pone.0033093.
Ghezzi D, Baruffini E, Haack TB, et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am J Hum Genet. 2012;90: 1079–87. https://doi.org/10.1016/j.ajhg.2012.04.011.
Baruffini E, Dallabona C, Invernizzi F, et al. MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum Mutat. 2013;34:1501–9. https://doi.org/10.1002/humu.22393.
Barry J Maron, MD, Julius M Gardin, MD, et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults echocardiographic Analysis of 4111 Subjects in the CARDIA Study. Circulation, 1995 ahajournals.org/content/92/4/785.
Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genom Hum Genet. 2010;11:25–44. https://doi.org/10.1146/annurev-genom-082509-141720.
Kohda M, Tokuzawa Y, Kishita Y, et al. A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet. 2016;12:e1005679. https://doi.org/10.1371/journal.pgen.1005679.
Nagao A, Suzuki T. Human mitochondrial tRNAs biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329. https://doi.org/10.1146/annurev-genet-110410-132531.
Echevarría L, Clemente P, Hernández-Sierra R, Gallardo ME, Fernández-Moreno MA. Glutamyl-tRNAGln amidotransferase is essential for mammalian mitochondrial translation in vivo. Biochem J. 2014;460:91–101. https://doi.org/10.1042/BJ20131107.
Nagao A, Suzuki T, Katoh T, Sakaguchi Y and Suzuki T. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc Natl Acad Sci USA. 2009;110.1073:16209–14 (received for review 9 July 2009).
Götz A, Tyynismaa H, Euro L, et al. Exome sequencing identifies mitochondrial Alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am J Hum Genet. 2011; 88:635-42.
Taylor RW, Pyle A, Griffin H, Blakely EL, Duff J. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA. 2014;312:68–77. https://doi.org/10.1001/jama.2014.7184.
Euro L, Konovalova S, Asin-Cayuela J, et al. Structural modeling of tissue-specific mitochondrial alanyl-tRNA synthetase (AARS2) defects predicts differential effects on aminoacylation. Front Genet. 2015;6:21. https://doi.org/10.3389/fgene.2015.00021.
Dallabona C, Diodato D, Kevelam SH, Haack TB, Wong LJ, Salomons GS. Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology. 2014;82:2063–71. https://doi.org/10.1212/WNL.0000000000000497.
Liu Y, Satz JS, Vo MN, Nangle LA, Schimmel P, Ackerman SL. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA. 2014;111:17570–5. https://doi.org/10.1073/pnas.1420196111.
Murphy E, Ardehali H, Balaban RS, American Heart Association Council on Basic Cardiovascular Sciences, Council on Clinical Cardiology, and Council on Functional Genomics and Translational Biology. Mitochondrial function, biology, and role in disease. A Scientific Statement From the American Heart Association. Circ Res. 2016;118:1960–91. https://doi.org/10.1161/RES.0000000000000104.
Coughlin CR 2nd, Scharer GH, Friederich MW, et al. Mutations in the mitochondrial cysteinyl-tRNA synthase gene CARS2, lead to a severe epileptic encephalopathy and complex movement disorder. J Med Genet. 2015;52:532–40. https://doi.org/10.1136/jmedgenet-2015-103049.
Taft RJ, Vanderver A, Leventer RJ, et al. Mutations in DARS cause hypomyelination with brain stem and spinal cord involvement and leg spasticity. Am J Hum Genet. 2013;92:774–80. https://doi.org/10.1016/j.ajhg.2013.04.006.
Steenweg ME, Ghezzi D, Haack T, et al. Leukoencephalopathy with thalamus and brain stem involvement and high lactate ‘LTBL’ caused by EARS2 mutations. Brain. 2012;135:1387–94. https://doi.org/10.1093/brain/aws070
Yadavalli SS, Euro L, et al. Mitochondrial phenylalanyl-tRNA synthetase mutations underlie fatal infabtile Alpers encephalopathy. Hum Mol Genet. 2012;21:4521–9.
Antonellis A1, Ellsworth RE, Sambuughin N, et al Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–9.
Pierce SB, Chisholm KM, Lynch ED, et al. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci USA. 2011;108:6543–8. https://doi.org/10.1073/pnas.1103471108.
Kopajtich R, Murayama K, Janecke AR, et al. Biallelic IARS mutations cause growth retardation with prenatal onset, intellectual disability, muscular hypotonia, and infantile hepatopathy. Am J Hum Genet. 2016;99:414–22. https://doi.org/10.1016/j.ajhg.2016.05.027.
Schwartzentruber Jl, Buhas D, Majewski J, et al. Mutation in the nuclear-encoded mitochondrial isoleucyl-tRNA synthetaseIARS2 in patients with cataracts, growth hormone deficiency with short stature, partial sensorineural deafness, and peripheral neuropathy or with Leigh syndrome. Hum Mutat. 2014;35:1285–9. https://doi.org/10.1002/humu.22629.
McLaughlin HM, Sakaguchi R, Liu C, NISC Comparative Sequencing Program, Biesecker LG, Wilson TE, Ionasescu V, Nicholson G, Searby C, Talbot K, Vance JM, Züchner S, Szigeti K, Lupski JR, Hou YM, Gree.n ED, Antonellis A. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet. 2010;87:560–6. https://doi.org/10.1016/j.ajhg.2010.09.008.
Pierce SB, Gersak K, Michaelson-Cohen R, et al. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am J Hum Genet. 2013;92:614–20. https://doi.org/10.1016/j.ajhg.2013.03.007.
Bayat V, Thiffault I, Jaiswal M, et al. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. https://doi.org/10.1371/journal.pbio.1001288.
Vanlander AV, Menten B, Smet J, et al. Two siblings with homozygous pathogenic splice-site variant in mitochondrial asparaginyl-tRNA synthetase (NARS2). Hum Mutat. 2015;36:222–31. https://doi.org/10.1002/humu.22728.
Simon M, Richard EM, Wang X, et al. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 2015;11:e1005097. https://doi.org/10.1371/journal.pgen.1005097.
Sofou K, Kollberg G, Holmström M, et al. Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol Genet Genom Med. 2015;3:59–68. https://doi.org/10.1002/mgg3.115.
Edvardson S, Shaag A, Kolesnikova O, et al. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet. 2007;81:857–62.
Belostotsky R, Ben-Shalom E, Rinat C, et al. Mutations in the mitochondrial seryl-tRNA synthetase cause hyperuricemia, pulmonary hypertension, renal failure in infancy and alkalosis, HUPRA syndrome. Am J Hum Genet. 2011;88:193–200. https://doi.org/10.1016/j.ajhg.2010.12.010.
Diodato D, Melchionda L, Haack TB, et al. VARS2 and TARS2 mutations in patients with mitochondrial encephalopathies. Hum Mutat. 2014;35:983–9. https://doi.org/10.1002/humu.22590.
Yan Q, Bykhovskaya Y, Li R, Mengesha E, et al. Human TRMU encoding the mitochondrial 5-methylaminomethyl-2-thiouridylate-methyltransferase is a putative nuclear modifier gene for the phenotypic expression of the deafness-associated 12S rRNA mutations. Biochem Biophys Res Commun. 2006;342:1130–6.
Zeharia A, Shaag A, Pappo O, et al. Acute infantile liver failure due to mutations in the TRMU gene. Am J Hum Genet. 2009;85:401–7. https://doi.org/10.1016/j.ajhg.2009.08.
Riley LG, Cooper S, Hickey P, et al. Mutations of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic aneamia-MLASA syndrome. Am J Hum Genet. 2010;87:52–9. https://doi.org/10.1016/j.ajhg.2010.06.001.
Acknowledgements
We acknowledge the patients and their families for their contribution to this study. Elvira N. Mulder-Den Hartog (Erasmus MC, Rotterdam) for clinical information, and Ralph W. Gottschalk (MUMC+, Maastricht) for the diagnostic analyses. This work was supported by a grant from the Princes Beatrix Spierfonds (W.OR11‐24) and Metakids.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures followed were in accordance with the ethical standards of the Helsinki Declaration of 1975, as revised in 2000.
Additional information
Jo M. Vanoevelen and Hubert J. M. Smeets contributed equally to this work.
Rights and permissions
About this article
Cite this article
Kamps, R., Szklarczyk, R., Theunissen, T.E. et al. Genetic defects in mtDNA-encoded protein translation cause pediatric, mitochondrial cardiomyopathy with early-onset brain disease. Eur J Hum Genet 26, 537–551 (2018). https://doi.org/10.1038/s41431-017-0058-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-017-0058-2
This article is cited by
-
Breastfeeding predicts blood mitochondrial DNA content in adolescents
Scientific Reports (2020)
-
Mitochondrial aminoacyl-tRNA synthetase disorders: an emerging group of developmental disorders of myelination
Journal of Neurodevelopmental Disorders (2019)
-
Recent topics: the diagnosis, molecular genesis, and treatment of mitochondrial diseases
Journal of Human Genetics (2019)
-
Pathogenic variants in glutamyl-tRNAGln amidotransferase subunits cause a lethal mitochondrial cardiomyopathy disorder
Nature Communications (2018)