Abstract
Background/objective
Although benefits of fish consumption for health are well known, a significant percentage of individuals dislike eating fish. Fish consumption may be influenced by genetic factors in addition to environmental factors. We conducted a genome-wide association study (GWAS) to find genetic variations that affect fish consumption in a Japanese population.
Methods
We performed a two-stage GWAS on fish consumption using 13,739 discovery samples from the Japan Multi-Institutional Collaborative Cohort study, and 2845 replication samples from the other population. We used a semi-quantitative food frequency questionnaire to estimate food intake. Association of the imputed variants with fish consumption was analyzed by separate linear regression models per variant, with adjustments for age, sex, energy intake, principal component analysis components 1–10, and alcohol intake (g/day). We also performed conditional analysis.
Results
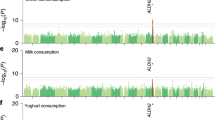
We found 27 single nucleotide polymorphisms (SNPs) located in 12q24 and 14q32.12 that were associated with fish consumption. The 19 SNPs were located at 11 genes including six lead SNPs at the BRAP, ACAD10, ALDH2, NAA25, and HECTD4 regions on 12q24.12-13, and CCDC197 region on 14q32.12. In replication samples, all five SNPs located on chromosome 12 were replicated successfully, but the one on chromosome 14 was not. Conditional analyses revealed that the five lead variants in chromosome 12 were in fact the same signal.
Conclusion
We found that new SNPs in the 12q24 locus were related to fish intake in two Japanese populations. The associations between SNPs on chromosome 12 and fish intake were strongly confounded by drinking status.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ascherio A, Rimm EB, Stampfer MJ, Giovannucci EL, Willett WC. Dietary intake of marine n-3 fatty acids, fish intake, and the risk of coronary disease among men. N Engl J Med. 1995;332:977–82.
Hu FB, Bronner L, Willett WC, Stampfer MJ, Rexrode KM, Albert CM, et al. Fish and omega-3 fatty acid intake and risk of coronary heart disease in women. JAMA. 2002;287:1815–21.
Albert CM, Campos H, Stampfer MJ, Ridker PM, Manson JE, Willett WC, et al. Blood levels of long-chain n-3 fatty acids and the risk of sudden death. N Engl J Med. 2002;346:1747–54.
Yamagishi K, Iso H, Date C, Fukui M, Wakai K, Kikuchi S, et al. Japan Collaborative Cohort Study for Evaluation of Cancer Risk Study Group. Fish, omega-3 polyunsaturated fatty acids, and mortality from cardiovascular diseases in a nationwide community-based cohort of Japanese men and women the JACC (Japan Collaborative Cohort Study for Evaluation of Cancer Risk) Study. J Am Coll Cardiol. 2008;52:988–96.
World Health Organization. European Food and Nutrition Action Plan 2015–2020. http://www.euro.who.int/__data/assets/pdf_file/0008/253727/64wd14e_FoodNutAP_140426.pdf.
Lloyd-Jones DM, Hong Y, Labarthe D, Mozaffarian D, Appel LJ, Van Horn L, et al. American Heart Association Strategic Planning Task Force and Statistics Committee. Defining and setting national goals for cardiovascular health promotion and disease reduction: the American Heart Association’s strategic Impact Goal through 2020 and beyond. Circulation. 2010;121:586–613.
Boesveldt S, Frasnelli J, Gordon AR, Lundström JN. The fish is bad: Negative food odors elicit faster and more accurate reactions than other odors. Biol Psychol. 2010;84:313–7.
Iso H, Sato S, Folsom AR, Shimamoto T, Terao A, Munger RG, et al. Serum fatty acids and fish intake in rural Japanese, urban Japanese, Japanese American and Caucasian American men. Int J Epidemiol. 1989;18:374–81.
Hirai A, Hamazaki T, Terano T, Nishikawa T, Tamura Y, Kamugai A, et al. Eicosapentaenoic acid and platelet function in Japanese. Lancet 1980;2:1132–3.
Kagawa Y, Nishizawa M, Suzuki M, Miyatake T, Hamamoto T, Goto K, et al. Eicosapolyenoic acids of serum lipids of Japanese islanders with low incidence of cardiovascular disease. J Nutr Sci Vitaminol. 1982;28:441–53.
Zhang J, Sasaki S, Amano K, Kesteloot H. Fish consumption and mortality from all causes, ischemic heart disease, and stroke: an ecological study. Prev Med 1999;28:520–9.
Nakamura Y, Ueshima H, Okamura T, Kadowaki T, Hayakawa T, Kita Y, et al. Association between fish consumption and all-cause and cause-specific mortality in Japan: NIPPON DATA80, 1980–99. Am J Med. 2005;118:239–45.
Mozaffarian D, Dashti HS, Wojczynski MK, Chu AY, Nettleton JA, Männistö S, et al. Genome-wide association meta-analysis of fish and EPA+DHA consumption in 17 US and European cohorts. PLoS One. 2017;12:e0186456.
Igarashi M, Nogawa S, Kawafune K, Hachiya T, Takahashi S, Jia H, et al. Identification of the 12q24 locus associated with fish intake frequency by genome-wide meta-analysis in Japanese populations. Genes Nutr 2019;14:21.
Hamajima N, J-MICC Study Group. The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for cancer. Asian Pac J Cancer Prev. 2007;8:317–23.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Yamaguchi-Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, Kubo M, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83:445–56.
1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Hamajima N, Matsuo K, Saito T, Hirose K, Inoue M, Takezaki T, et al. Gene-environment Interactions and Polymorphism Studies of Cancer Risk in the Hospital-based Epidemiologic Research Program at Aichi Cancer Center II (HERPACC-II). Asian Pac J Cancer Prev. 2001;2:99–107.
Imaeda N, Fujiwara N, Tokudome Y, Ikeda M, Kuriki K, Nagaya T, et al. Reproducibility of a semi-quantitative food frequency questionnaire in Japanese female dietitians. J Epidemiol. 2002;12:45–53.
Goto C, Tokudome Y, Imaeda N, Takekuma K, Kuriki K, Igarashi F, et al. Validation study of fatty acid consumption assessed with a short food frequency questionnaire against plasma concentration in middle-aged Japanese. Scand J Nutr. 2006;2:77–82.
Tokudome Y, Goto C, Imaeda N, Hasegawa T, Kato R, Hirose K, et al. Relative validity of a short food frequency questionnaire for assessing nutrient intake versus three-day weighed diet records in middle-aged Japanese. J Epidemiol. 2005;15:135–45.
Tokudome S, Goto C, Imaeda N, Tokudome Y, Ikeda M, Mizuno M. Development of a data-based short food frequency questionnaire for assessing nutrient intake by middle-aged Japanese. Asian Pac J Cancer Prev. 2004;5:40–3.
Delaneau O, Marchini J, Zagury J. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–81.
Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7.
Das S. DosageConvertor. https://genome.sph.umich.edu/wiki/DosageConvertor.
Delongchamp R, Faramawi MF, Feingold E, Chung D, Abouelenein S. The Association between SNPs and a Quantitative Trait: Power Calculation. Eur J Environ Public Health. 2018;2:1–7.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Willett WC, Howe GR, Kushi LH. Adjustment for total energy intake in epidemiologic studies. Am J Clin Nutr. 1997;65(4 Suppl):1220S–8S.
Zhu X, Li S, Cooper RS, Elston RC. A unified association analysis approach for family and unrelated samples correcting for stratifications. Am J Hum Genet. 2008;82:352–65.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Gogarten SM, Bhangale T, Conomos MP, Laurie CA, McHugh CP, Painter I, et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–31.
Turner, SD. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. biorXiv 2014. https://doi.org/10.1101/005165.
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7.
Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinforma 2010;11:288.
da Cunha Veloso MC, da Silva VM, Santos GV, de Andrade JB. Determination of aldehydes in fish by high-performance liquid chromatography. J Chromatogr Sci. 2001;39:173–6.
Uebelacker M, Lachenmeier DW. Quantitative determination of acetaldehyde in foods using automated digestion with simulated gastric fluid followed by headspace gas chromatography. J Autom Methods Manag Chem. 2011;2011:907317.
Kosowka M, Majcher MA, Fortuna T. Volatile compound in meat and meat products. Food Sci Technol Camp. 2017;3:1–7.
Okada Y, Momozawa Y, Sakaue S, Kanai M, Ishigaki K, Akiyama M, et al. Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat Commun. 2018;9:1631.
Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478:103–9.
Acknowledgements
We would like to thank all the staff at the Laboratory for Genotyping Development, Center for the Integrative Medical Sciences, RIKEN, and the staff of the BioBank Japan project. This study was supported by a Grants-in-Aid for Scientific Research for Priority Areas of Cancer (No. 17015018) and Innovative Areas (No. 221S0001), and by JSPS KAKENHI Grants (No. 16H06277) from the Japanese Ministry of Education, Culture, Sports, Science and Technology. This work was also supported in part by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant B Numbers 24390165, 20390184, 17390186. This study was supported in part by funding for the BioBank Japan Project from the Japan Agency for Medical Research and development from April 2015, and the Ministry of Education, Culture, Sports, Science and Technology from April 2003 to March 2015.
J-MICC Research Group
Kenji Wakai22, Kenji Takeuchi22, Haruo Mikami19, Hiroki Nagase25, Hiroto Narimatsu26,27, Kiyonori Kuriki12, Sadao Suzuki20, Keitaro Matsuo4, Asahi Hishida22, Yoshikuni Kita2,24, Katsuyuki Miura2,28, Ritei Uehara18, Kokichi Arisawa21, Hiroaki Ikezaki14, Keitaro Tanaka13, Toshiro Takezaki16.
Author information
Authors and Affiliations
Consortia
Contributions
TS, YaN, YoKi, and KW: designed the research; KeMa, KaMi, NT, KK, CS, KeTa, HiIk, MM, RI, ToT, YuK, HiIt, DM, TK, HM, YoN, SS, TN, SKK, KA, KenT, TaTa, RO, YoKu, YM, MK, YoKi, and KW: conducted the research; TS, YaN, KeMa, IO, AN, AS, NI, CG, and MN: analyzed data and performed statistical analysis; TS, YaN, KeMa, IO, and YD: wrote the manuscript and had primary responsibility for final content; and all authors: read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Suzuki, T., Nakamura, Y., Matsuo, K. et al. A genome-wide association study on fish consumption in a Japanese population—the Japan Multi-Institutional Collaborative Cohort study. Eur J Clin Nutr 75, 480–488 (2021). https://doi.org/10.1038/s41430-020-00702-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41430-020-00702-7
This article is cited by
-
A genome-wide association study on adherence to low-carbohydrate diets in Japanese
European Journal of Clinical Nutrition (2022)