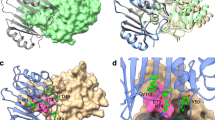
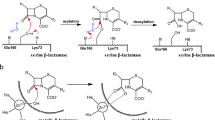
Abstract
Dissemination of class D OXA-type carbapenemases is one of the significant causes of beta-lactam resistance in Gram-negative bacteria. The amino acid residues present near the active site are involved in hydrolytic mechanism of class D carbapenemases, though it is not identified in OXA-23. Here, with the help of site-directed mutagenesis, we aimed to explicate the importance of the residues W165, L166 and V167 of the possible omega loop and residue D222 in the short β5-β6 loop on the activity of OXA-23. All the residues were substituted with alanine. The resultant proteins were assayed for the changes in activity in E. coli cells and purified for in vitro activity, and stability assessment. E. coli cells harboring OXA-23_W165A and OXA-23_L166A, individually, exhibited a significant decrease in resistance towards beta-lactam antibiotics as compared to OXA-23. Further, purified OXA-23_W165A and OXA-23_L166A imparted about >4-fold decrease in catalytic efficiency and displayed reduced thermal stability as compared to OXA-23. Bocillin-FL binding assay revealed that W165A substitution results in improper N-carboxylation of K82, leading to deacylation deficient OXA-23. Therefore, we infer that the residue W165 maintains the integrity of N-carboxylated lysine (K82) of OXA-23 and the residue L166 might be responsible for properly orientating the antibiotic molecules.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–31.
Vuillemenot JB, Bour M, Beyrouthy R, Bonnet R, Laaberki MH, Charpentier X, et al. Genomic analysis of CTX-M-115 and OXA-23/-72 co-producing Acinetobacter baumannii, and their potential to spread resistance genes by natural transformation. J Antimicrob Chemother. 2022;77:1542–52.
Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of bla(OXA-23) with bla(NDM-1) and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother. 2010;65:2253–4.
La MV, Jureen R, Lin RTP, Teo JWP. Unusual detection of an acinetobacter class D carbapenemase gene, bla(OXA-23), in a clinical Escherichia coli isolate. J Clin Microbiol. 2014;52:3822–3.
Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–76.
Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54:24–38.
Walther-Rasmussen J, Hoiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57:373–83.
Donald HM, Scaife W, Amyes SG, Young HK. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44:196–9.
Héritier C, Poirel L, Lambert T, Nordmann P. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49:3198–202.
Lee Y, Yum JH, Kim CK, Yong D, Jeon EH, Jeong SH, et al. Role of OXA-23 and AdeABC efflux pump for acquiring carbapenem resistance in an Acinetobacter baumannii strain carrying the blaOXA-66 gene. Ann Clin Lab Sci. 2010;40:43–48.
Evans BA, Amyes SG. OXA beta-lactamases. Clin Microbiol Rev. 2014;27:241–63.
Rasmussen BA, Bush K. Carbapenem-hydrolyzing beta-lactamases. Antimicrob Agents Chemother. 1997;41:223–32.
Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases. Proc Natl Acad Sci USA. 2001;98:14280–5.
Verma V, Testero SA, Amini K, Wei W, Liu J, Balachandran N, et al. Hydrolytic mechanism of OXA-58 enzyme, a carbapenem-hydrolyzing class D beta-lactamase from Acinetobacter baumannii. J Biol Chem. 2011;286:37292–303.
Jain D, Verma J, Ghosh AS. Deciphering the role of residues in the loops nearing the active site of OXA-58 in imparting beta-lactamase activity. Microbiology. 2022;168:001203.
Hocquet D, Colomb M, Dehecq B, Belmonte O, Courvalin P, Plesiat P, et al. Ceftazidime-hydrolysing beta-lactamase OXA-145 with impaired hydrolysis of penicillins in Pseudomonas aeruginosa. J Antimicrob Chemother. 2011;66:1745–50.
Paetzel M, Danel F, de Castro L, Mosimann SC, Page MG, Strynadka NC. Crystal structure of the class D beta-lactamase OXA-10. Nat Struct Biol. 2000;7:918–25.
Stewart NK, Toth M, Alqurafi MA, Chai W, Nguyen TQ, Quan P, et al. C6 hydroxymethyl-substituted carbapenem MA-1-206 inhibits the major Acinetobacter baumannii carbapenemase OXA-23 by impeding deacylation. mBio. 2022;13:e0036722.
Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6.
Wayne P. Clinical Laboratory Standard Institute. CLSI performance standard of antimicrobial susceptibility testing: twenty-fourth international supplement. CLSI Doc M100-S24. 2014;34:50–106.
Verma J, Jain D, Mallik D, Ghosh AS. Comparative insight into the roles of the non active-site residues E169 and N173 in imparting the beta-lactamase activity of CTX-M-15. Fems Microbiol Lett. 2022;369:fnac018.
Kaitany KC, Klinger NV, June CM, Ramey ME, Bonomo RA, Powers RA, et al. Structures of the class D Carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam. Antimicrob Agents Chemother. 2013;57:4848–55.
Baurin S, Vercheval L, Bouillenne F, Falzone C, Brans A, Jacquamet L, et al. Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases. Biochemistry. 2009;48:11252–63.
Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43:1124–8.
Bansal A, Kar D, Murugan RA, Mallick S, Dutta M, Pandey SD, et al. A putative low-molecular-mass penicillin-binding protein (PBP) of Mycobacterium smegmatis exhibits prominent physiological characteristics of DD-carboxypeptidase and beta-lactamase. Microbiology. 2015;161:1081–91.
Pratap S, Katiki M, Gill P, Kumar P, Golemi-Kotra D. Active-site plasticity is essential to carbapenem hydrolysis by OXA-58 class D β-lactamase of Acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60:75–86.
Santillana E, Beceiro A, Bou G, Romero A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc Natl Acad Sci USA. 2007;104:5354–9.
Kumar G, Issa B, Kar D, Biswal S, Ghosh AS. E152A substitution drastically affects NDM-5 activity. FEMS Microbiol Lett 2017; 364:fnx008.
Kumar G, Biswal S, Nathan S, Ghosh AS. Glutamate residues at positions 162 and 164 influence the beta-lactamase activity of SHV-14 obtained from Klebsiella pneumoniae. Fems Microbiol Lett. 2018;365:fnx259.
Kumar G, Issa B, Biswal S, Jain D, Bhattacharjee A, Ghosh AS. Glutamic acid at position 152 and serine at position 191 are key residues required for the metallo-β-lactamase activity of NDM-7. Int J Antimicrob Agents. 2020;55:105824.
Smith CA, Antunes NT, Stewart NK, Toth M, Kumarasiri M, Chang M, et al. Structural basis for carbapenemase activity of the OXA-23 beta-lactamase from Acinetobacter baumannii. Chem Biol. 2013;20:1107–15.
Smith CA, Stewart NK, Toth M, Vakulenko SB. Structural insights into the mechanism of carbapenemase activity of the OXA-48 β-lactamase. Antimicrob Agents Chemother. 2019;63:e01202–01219.
Mitchell JM, Clasman JR, June CM, Kaitany KC, LaFleur JR, Taracila MA, et al. Structural basis of activity against aztreonam and extended spectrum cephalosporins for two carbapenem-hydrolyzing class D beta-lactamases from Acinetobacter baumannii. Biochemistry. 2015;54:1976–87.
Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95.
Schneider KD, Bethel CR, Distler AM, Hujer AM, Bonomo RA, Leonard DA. Mutation of the active site carboxy-lysine (K70) of OXA-1 beta-lactamase results in a deacylation-deficient enzyme. Biochemistry. 2009;48:6136–45.
Adachi H, Ohta T, Matsuzawa H. Site-directed mutants, at position 166, of Rtem-1 beta-lactamase that form a stable acyl-enzyme intermediate with penicillin. J Biol Chem. 1991;266:3186–91.
Funding
ASG thanks the Department of Biotechnology (DBT), Government of India [Grant # BT/PR24255/NER/95/716/2017] and Council of Scientific and Industrial Research (CSIR) [Grant # 27(0367)/20/EMR-II] for the research support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jain, D., Verma, J., Ajith, T. et al. Two non-active site residues W165 and L166 prominently influence the beta-lactam hydrolytic ability of OXA-23 beta-lactamase. J Antibiot 76, 489–498 (2023). https://doi.org/10.1038/s41429-023-00624-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-023-00624-z