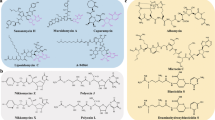
Abstract
Thiopeptides are a class of natural product antibiotics with diverse structures and functions. Their complex structures and biosynthesis have intrigued researchers since their discovery in 1948, but not a single thiopeptide has been approved for human use. This is mainly due to their poor solubility, challenging synthesis, and low bioavailability. This review summarizes the current research on the biosynthesis and biological activity of thiopeptide antibiotics since 2015. The focus of research since 2015 has been on uncovering biosynthetic routes, developing methods for total synthesis, and understanding the biological activity of thiopeptides. Overall, there is still much to learn about this family of molecules.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Liu G, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev. 2013;77:112–43.
Qin Z, Munnoch JT, Devine R, Holmes NA, Seipke RF, Wilkinson KA, et al. Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem Sci. 2017;8:3218–27.
Seipke RF, Kaltenpoth M, Hutchings MI. Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev. 2012;36:862–76.
Hu X, Hu X, Hu X, Li S, Li L, Yu L, et al. Cytotoxic and antibacterial cervinomycins B1-4 from a Streptomyces species. J Nat Prod. 2019;82:2337–42.
de Lima Procópio RE, da Silva IR, Martins MK, de Azevedo JL, de Araújo JM. Antibiotics produced by Streptomyces. Braz J Infect Dis. 2012;16:466–71.
Linares JF, Gustafsson I, Baquero F, Martinez JL. Antibiotics as intermicrobiol signaling agents instead of weapons. Proc Natl Acad Sci USA. 2006;103:19484–9.
Romero D, Traxler MF, López D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–505.
Li S, Hu X, Li L, Liu H, Yu L, You X, et al. Geninthiocins C and D from Streptomyces as 35-membered macrocyclic thiopeptides with modified tail moiety. J Antibiot. 2019;72:106–10.
Schneider O, Simic N, Aachmann FL, Rückert C, Kristiansen KA, Kalinowski J, et al. Genome mining of Streptomyces sp. YIM 130001 isolated from lichen affords new thiopeptide antibiotic. Front Microbiol. 2018;9:3139.
Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–60.
Su TL. Micrococcin, an antibacterial substance formed by a strain of Micrococcus. Br J Exp Pathol. 1948;29:473–81.
Li J, Qu X, He X, Duan L, Wu G, Bi D, et al. ThioFinder: a web-based tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS ONE. 2012;7:e45878.
Schwalen CJ, Hudson GA, Kille B, Mitchell DA. Bioinformatic expansion and discovery of thiopeptide antibiotics. J Am Chem Soc. 2018;140:9494–501.
Little R, Paiva FCR, Jenkins R, Hong H, Sun Y, Demydchuk Y, et al. Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis. Nat Catal. 2019;2:1045–54.
Cogan DP, Hudson GA, Zhang Z, Pogorelov TV, Van Der Donk WA, Mitchell DA, et al. Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis. Proc Natl Acad Sci USA. 2017;114:12928–33.
Fleming SR, Bartges TE, Vinogradov AA, Kirkpatrick CL, Goto Y, Suga H, et al. Flexizyme-enabled benchtop biosynthesis of thiopeptides. J Am Chem Soc. 2019;141:758–62.
Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714.
Khilyas IV, Tursunov KA, Shirshikova TV, Kamaletdinova LK, Matrosova LE, Desai PT, et al. Genome sequence of pigmented siderophore-producing strain Serratia marcescens SM6. Microbiol Resour Announc. 2019;8:e00247–19.
Cheng TH, Saidin J, Danish-Daniel M, Gan HM, Isa MNM, Bakar MFA, et al. Genome sequence of Serratia marcescens subsp. sakuensis strain K27, a marine bacterium isolated from sponge (Haliclona amboinensis). Genome Announc. 2018;6:e00022–18.
Thompson J, Cundliffe E, Stark MJR. The mode of action of berninamycin and the mechanism of resistance in the producing organism, Streptomyces bernensis. J Gen Microbiol. 1982;128:875–84.
Naaktgeboren N, Roobol K, Gubbens J, Voorma HO. The mode of action of thiostrepton in the initiation of protein synthesis. Eur J Biochem. 1976;70:39–47.
Cundliffe E, Thompson J. The mode of action of nosiheptide (multhiomycin) and the mechanism of resistance in the producing organism. J Gen Microbiol. 1981;126:185–92.
Polikanov YS, Aleksashin NA, Beckert B, Wilson DN. The mechanisms of action of ribosome-targeting peptide antibiotics. Front Mol Biosci. 2018;5:48.
Walter JD, Hunter M, Cobb M, Traeger G, Spiegel PC. Thiostrepton inhibits stable 70S ribosome binding and ribosome-dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Res. 2012;40:360–70.
Bowen WS, Van Dyke N, Murgola EJ, Lodmell JS, Hill WE. Interaction of thiostrepton and elongation factor-G with the ribosomal protein L11-binding domain. J Biol Chem. 2005;280:2934–43.
Highland JH, Lin L, Bodley JW. Protection of ribosomes from thiostrepton inactivation by the binding of G factor and guanosine diphosphate. Biochemistry. 1971;10:4404–9.
Bodley JW, Lin L, Highland JH. Studies on translocation VI: thiostrepton prevents the formation of a ribosome-G factor-guanine nucleotide complex. Biochem Biophys Res Commun. 1970;41:1406–11.
Rodnina MV, Savelsbergh A, Matassova NB, Katunin VI, Semenkov YP, Wintermeyer W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc Natl Acad Sci USA. 1999;96:9586–90.
Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, et al. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30:26–38.
Bechthold A, Floss HG. Overexpression of the thiostrepton‐resistance gene from Streptomyces azureus in Escherichia coli and characterization of recognition sites of the 23S rRNA A1067 2′‐methyltransferase in the guanosine triphosphatase center of 23S ribosomal RNA. Eur J Biochem. 1994;224:431–7.
Thompson J, Schmidt F, Cundliffe E. Site of action of a ribosomal RNA methylase conferring resistance to thiostrepton. J Biol Chem. 1982;257:7915–7.
Yin S, Jiang H, Chen D, Murchie AIH. Substrate recognition and modification by the nosiheptide resistance methyltransferase. PLoS ONE. 2015;10:e0122972.
Rosendahl G, Douthwaite S. The antibiotics micrococcin and thiostrepton interact directly with 23S rRNA nucleotides 1067A and 1095A. Nucleic Acids Res. 1994;22:357–63.
Lentzen G, Klinck R, Matassova N, Aboul-Ela F, Murchie AIH. Structural basis for contrasting activities of ribosome binding thiazole antibiotics. Chem Biol. 2003;10:769–78.
Parmeggiani A, Krab IM, Okamura S, Nielsen RC, Nyborg J, Nissen P. Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu. Biochemistry. 2006;45:6846–57.
Zuurmond AM, De Graaf JM, Olsthoorn-Tieleman LN, Van Duyl BY, Mörhle VG, Jurnak F, et al. Ge2270A-resistant mutations in elongation factor Tu allow productive aminoacyl-tRNA binding to EF-TU·GTP·GE2270A complexes. J Mol Biol. 2000;304:995–1005.
Young TS, Dorrestein PC, Walsh CT. Codon randomization for rapid exploration of chemical space in thiopeptide antibiotic variants. Chem Biol. 2012;19:1600–10.
Hashimoto M, Murakami T, Funahashi K, Tokunaga T, Nihei KI, Okuno T, et al. An RNA polymerase inhibitor, cyclothiazomycin B1, and its isomer. Bioorg Med Chem. 2006;14:8259–70.
Mizuhara N, Kuroda M, Ogita A, Tanaka T, Usuki Y, Fujita KI. Antifungal thiopeptide cyclothiazomycin B1 exhibits growth inhibition accompanying morphological changes via binding to fungal cell wall chitin. Bioorganic. Med Chem. 2011;19:5300–10.
Hayashi S, Ozaki T, Asamizu S, Ikeda H, Omura S, Oku N, et al. Genome mining reveals a minimum gene set for the biosynthesis of 32-membered macrocyclic thiopeptides lactazoles. Chem Biol. 2014;21:679–88.
Vinogradov AA, Shimomura M, Goto Y, Ozaki T, Asamizu S, Sugai Y, et al. Minimal lactazole scaffold for in vitro thiopeptide bioengineering. Nat Commun. 2020;11:1–13.
Vinogradov AA, Shimomura M, Kano N, Goto Y, Onaka H, Suga H. Promiscuous enzymes cooperate at the substrate level en route to lactazole A. J Am Chem Soc. 2020;142:13886–97.
Kongsema M, Wongkhieo S, Khongkow M, Lam EWF, Boonnoy P, Vongsangnak W, et al. Molecular mechanism of forkhead box M1 inhibition by thiostrepton in breast cancer cells. Oncol Rep. 2019;42:953–62.
Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem. 2011;3:725–31.
Halasi M, Gartel AL. A novel mode of FoxM1 regulation: positive auto-regulatory loop. Cell Cycle. 2009;8:1966–7.
Nicolaou KC, Zak M, Rahimipour S, Estrada AA, Lee SH, O’Brate A, et al. Discovery of a biologically active thiostrepton fragment. J Am Chem Soc. 2005;127:15042–4.
Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS ONE. 2009;4:e5592.
Aminake MN, Schoof S, Sologub L, Leubner M, Kirschner M, Arndt HD, et al. Thiostrepton and derivatives exhibit antimalarial and gametocytocidal activity by dually targeting parasite proteasome and apicoplast. Antimicrob Agents Chemother. 2011;55:1338–48.
Goodman CD, Su V, McFadden GI. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2007;152:181–91.
McConkey GA, Rogers MJ, McCutchan TF. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J Biol Chem. 1997;272:2046–9.
Bousema T, Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin Microbiol Rev. 2011;24:377–410.
Lucantoni L, Silvestrini F, Signore M, Siciliano G, Eldering M, Dechering KJ, et al. A simple and predictive phenotypic high content imaging assay for Plasmodium falciparum mature gametocytes to identify malaria transmission blocking compounds. Sci Rep. 2015;5:1–14.
Mugweru J, Makafe G, Cao Y, Zhang Y, Wang B, Huang S, et al. A cassette containing thiostrepton, gentamicin resistance genes, and dif sequences is effective in construction of recombinant Mycobacteria. Front Microbiol. 2017;8:468.
Fouces R, RodrÃguez M, Mellado E, DÃez B, Barredo JL. Conjugation and transformation of Streptomyces species by tylosin resistance. FEMS Microbiol Lett. 2006;186:319–25.
Chiu ML, Folcher M, Griffin P, Holt T, Klatt T, Thompson CJ. Characterization of the covalent binding of thiostrepton to a thiostrepton-induced protein from Streptomyces lividans. Biochemistry. 1996;35:2332–41.
Murakami T, Holt TG, Thompson CJ. Thiostrepton-induced gene expression in Streptomyces lividans. J Bacteriol. 1989;171:1459–66.
Chiu ML, Viollier PH, Katoh T, Ramsden JJ, Thompson CJ. Ligand-induced changes in the Streptomyces lividans TipAL protein imply an alternative mechanism of transcriptional activation for MerR-like proteins. Biochemistry. 2001;40:12950–8.
Holmes DJ, Caso JL, Thompson CJ. Autogenous transcriptional activation of a thiostrepton induced gene in Streptomyces lividans. EMBO J. 1993;12:3183–91.
Chiu ML, Folcher M, Katoh T, Puglia AM, Vohradsky J, Yun BS, et al. Broad spectrum thiopeptide recognition specificity of the Streptomyces lividans TipAL protein and its role in regulating gene expression. J Biol Chem. 1999;274:20578–86.
Myers CL, Harris J, Yeung JCK, Honek JF. Molecular interactions between thiostrepton and the TipAS protein from Streptomyces lividans. ChemBioChem. 2014;15:681–7.
Yun BS, Hidaka T, Furihata K, Seto H. Promothiocins A and B, novel thiopeptides with a tip A promoter inducing activity produced by Streptomyces sp. SF2741. J Antibiot. 1994;47:510–4.
González Holgado G, Castro Rodríguez J, Cañedo Hernandez LM, Díaz M, Fernández-Abalos JM, Trujillano I, et al. Radamycin, a novel thiopeptide produced by Streptomyces sp. RSP9. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot. 2002;55:383–99.
Bleich R, Watrous JD, Dorrestein PC, Bowers AA, Shank EA. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc Natl Acad Sci USA. 2015;112:3086–91.
Gotoh H, Zhang Y, Dallo SF, Hong S, Kasaraneni N, Weitao T. Pseudomonas aeruginosa, under DNA replication inhibition, tends to form biofilms via Arr. Res Microbiol. 2008;159:294–302.
Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–5.
Jin Y, Guo Y, Zhan Q, Shang Y, Qu D, Yu F. Subinhibitory concentrations of mupirocin stimulate Staphylococcus aureus biofilm formation by upregulating cidA. Antimicrob Agents Chemother. 2020;64:e01912–19.
Szczuka E, Jabłońska L, Kaznowski A. Effect of subinhibitory concentrations of tigecycline and ciprofloxacin on the expression of biofilm-associated genes and biofilm structure of Staphylococcus epidermidis. Microbiology. 2017;163:712–8.
Ranieri MRM, Chan DCK, Yaeger LN, Rudolph M, Karabelas-Pittman S, Abdo H, et al. Thiostrepton hijacks pyoverdine receptors to inhibit growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63:e00472–19.
Fougera Pharmaceuticals Inc (for Dechra). ANIMAX® Ointment (nystatin-neomycin sulfate-thiostrepton-triamcinolone acetonide) safety data sheet. Fougera Pharmaceuticals Inc; United States 2015.
Benazet F, Cartier JR. Effect of nosiheptide as a feed additive in chicks on the quantity, duration, prevalence of excretion, and resistance to antibacterial agents of Salmonella typhimurium; on the proportion of Escherichia coli and other coliforms resistant to antibacterial agents; and on their degree and spectrum of resistance. Poult Sci. 1980;59:1405–15.
Pascal C, Gaillard C, Moreau M-O. Identification of nosiheptide in feeds and detection of residues in animal tissues. J AOAC Int. 1979;62:976–81.
Song X, Xie J, Su Y, Martín-Esteban A, Qiu J, Li X, et al. Analysis of nosiheptide in food animal tissues via its unique degradation product by liquid chromatography-tandem mass spectrometry after alkaline hydrolysis. J Agric Food Chem. 2019;67:10791–9.
The Japan Food chemical Research Faundation. MRLs list: compositional specification for foods. The Ministry of Health, Labour and Welfare; 2017. https://www.ffcr.or.jp/en/zanryu/the-japanese-positive/the-japanese-positive-list-system-for-agricultural-chemical-residues-in-foods-enforcement-on-may-29-.html. Accessed 12 Apr 2020. Updated on December 25, 2017.
European Union. Commission regulation (EU) No 37/2010 on pharmacologically active substances and their classification regarding maximum residue limits in foodstuffs of animal origin. EUR-LEX; 2010. Accessed 12 Apr 2020. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:02010R0037-20200209.
Lassalas P, Berini C, Rouchet JBEY, Hédouin J, Marsais F, Schneider C, et al. Miyaura borylation/Suzuki-Miyaura coupling (MBSC) sequence of 4-bromo-2,4′-bithiazoles with halides: straightforward access to a heterocylic cluster of d-series of thiopeptide GE2270. Org Biomol Chem. 2018;16:526–30.
Citron DM, Tyrrell KL, Merriam CV, Goldstein EJC. Comparative in vitro activities of LFF571 against Clostridium difficile and 630 other intestinal strains of aerobic and anaerobic bacteria. Antimicrob Agents Chemother. 2012;56:2493–503.
Mullane K, Lee C, Bressler A, Buitrago M, Weiss K, Dabovic K, et al. Multicenter, randomized clinical trial to compare the safety and efficacy of lff571 and vancomycin for Clostridium difficile infections. Antimicrob Agents Chemother. 2015;59:1435–40.
ClinicalTrials.gov. Safety and efficacy of multiple daily dosing of oral LFF571 in patients with moderate Clostridium difficile infections. ClinicalTrials.gov; 2016. https://clinicaltrials.gov/ct2/show/NCT01232595. Accessed 6 Apr 2020. Novartis Pharmaceuticals. Identifier: NCT01232595.
Bhansali SG, Mullane K, Ting LSL, Leeds JA, Dabovic K, Praestgaard J, et al. Pharmacokinetics of LFF571 and vancomycin in patients with moderate Clostridium difficile infections. Antimicrob Agents Chemother. 2015;59:1441–5.
Kasumov A. Novartis Exits Antibiotics Research, cuts 140 jobs in Bay Area. Bloomberg; 2018. https://www.bnnbloomberg.ca/novartis-exits-antibiotics-research-cuts-140-jobs-in-bay-area-1.1106587. Accessed 6 Apr 2020.
NAICONS Srl. Clinical efficacy and safety of NAI-Acne gel 3% applied twice-a-day to patients with facial acne vulgaris. https://www.clinicaltrialsregister.eu/ctr-search/search?query=2014-001491-62. European Medicines Agency: EU Clinical Trials Register: Slovakia. Accessed 6 Apr 2020.
Cassiopea SPA. Interim report as of and for the three months ended March 31, 2020. Cassiopea SPA; 2020. https://www.cassiopea.com/wp-content/uploads/2020/05/CASSIOPEA-CONSO-Q1-2020.pdf. Accessed 6 Apr 2020.
Fabbretti A, He CG, Gaspari E, Maffioli S, Brandi L, Spurio R, et al. A derivative of the thiopeptide GE2270A highly selective against Propionibacterium acnes. Antimicrob Agents Chemother. 2015;59:4560–8.
Jin L, Wu X, Xue Y, Jin Y, Wang S, Chen Y. Mutagenesis of NosM leader peptide reveals important elements in nosiheptide biosynthesis. Appl Environ Microbiol. 2017;83:e02880–16.
Engelhardt K, Degnes KF, Zotchev SB. Isolation and characterization of the gene cluster for biosynthesis of the thiopeptide antibiotic TP-1161. Appl Environ Microbiol. 2010;76:7093–101.
Yu Y, Duan L, Zhang Q, Liao R, Ding Y, Pan H, et al. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem Biol. 2009;4:855–64.
Zhang Z, Hudson GA, Mahanta N, Tietz JI, Van der Donk WA, Mitchell DA. Biosynthetic timing and substrate specificity for the thiopeptide thiomuracin. J Am Chem Soc. 2016;138:15511–4.
Wang B, Lamattina JW, Marshall SL, Booker SJ. Capturing intermediates in the reaction catalyzed by NosN, a class C radical S-adenosylmethionine methylase involved in the biosynthesis of the nosiheptide side-ring system. J Am Chem Soc. 2019;141:5788–97.
Amara P, Mouesca JM, Bella M, Martin L, Saragaglia C, Gambarelli S, et al. Radical S-adenosyl-l-methionine tryptophan lyase (NosL): how the protein controls the carboxyl radical CO2− migration. J Am Chem Soc. 2018;140:16661–8.
Ji X, Li Y, Ding W, Zhang Q. Substrate-tuned catalysis of the radical S-adenosyl-L-methionine enzyme NosL involved in nosiheptide biosynthesis. Angew Chem Int Ed Engl. 2015;54:9021–4.
Badding ED, Grove TL, Gadsby LK, Lamattina JW, Boal AK, Booker SJ. Rerouting the pathway for the biosynthesis of the side ring system of nosiheptide: the roles of NosI, NosJ, and NosK. J Am Chem Soc. 2017;139:5896–905.
Ding W, Ji W, Wu Y, Wu R, Liu WQ, Mo T, et al. Biosynthesis of the nosiheptide indole side ring centers on a cryptic carrier protein NosJ. Nat Commun. 2017;437:8.
Ding W, Li Y, Zhao J, Ji X, Mo T, Qianzhu H, et al. The catalytic mechanism of the class C radical S-adenosylmethionine methyltransferase NosN. Angew Chem Int Ed Engl. 2017;56:3857–61.
Ding W, Wu Y, Ji X, Qianzhu H, Chen F, Deng Z, et al. Nucleoside-linked shunt products in the reaction catalyzed by the class C radical S-adenosylmethionine methyltransferase NosN. Chem Commun. 2017;53:5235–8.
LaMattina JW, Wang B, Badding ED, Gadsby LK, Grove TL, Booker SJ. NosN, a radical S-adenosylmethionine methylase, catalyzes both C1 transfer and formation of the ester linkage of the side-ring system during the biosynthesis of nosiheptide. J Am Chem Soc. 2017;139:17438–45.
Qiu Y, Du Y, Wang S, Zhou S, Guo Y, Liu W. Radical S-adenosylmethionine protein NosN forms the side ring system of nosiheptide by functionalizing the polythiazolyl peptide S-conjugated indolic moiety. Org Lett. 2019;21:1502–5.
Wang Y, Liu S, Yao P, Yu Y, Zhang Y, Lan W, et al. Crystallographic analysis of NosA, which catalyzes terminal amide formation in the biosynthesis of nosiheptide. Acta Crystallogr Sect Struct Biol Commun. 2015;71:1033–7.
Yu Y, Guo H, Zhang Q, Duan L, Ding Y, Liao R, et al. NosA catalyzing carboxyl-terminal amide formation in nosiheptide maturation via an enamine dealkylation on the serine-extended precursor peptide. J Am Chem Soc. 2010;132:16324–6.
Wu X, Jin L, Zhang H, Tong R, Ma M, Chen Y. Identification of truncated form of NosP as a transcription factor to regulate the biosynthesis of nosiheptide. FASEB J. 2018;32:453–65.
Hudson GA, Zhang Z, Tietz JI, Mitchell DA, Van Der Donk WA. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J Am Chem Soc. 2015;137:16012–5.
Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16:141–7.
Lin Z, Ji J, Zhou S, Zhang F, Wu J, Guo Y, et al. Processing 2-methyl-l-tryptophan through tandem transamination and selective oxygenation initiates indole ring expansion in the biosynthesis of thiostrepton. J Am Chem Soc. 2017;139:12105–8.
Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–34.
Zhang F, Kelly WL. Saturation mutagenesis of TsrA Ala4 unveils a highly mutable residue of thiostrepton A. ACS Chem Biol. 2015;10:998–1009.
Duan L, Wang S, Liao R, Liu W. Insights into quinaldic acid moiety formation in thiostrepton biosynthesis facilitating fluorinated thiopeptide generation. Chem Biol. 2012;19:443–8.
Zheng Q, Wang S, Duan P, Liao R, Chen D, Liu W. An α/β-hydrolase fold protein in the biosynthesis of thiostrepton exhibits a dual activity for endopeptidyl hydrolysis and epoxide ring opening/macrocyclization. Proc Natl Acad Sci USA. 2016;113:14318–23.
Luo X, Zambaldo C, Liu T, Zhang Y, Xuan W, Wang C, et al. Recombinant thiopeptides containing noncanonical amino acids. Proc Natl Acad Sci USA. 2016;113:3615–20.
Chan DCK, Guo I, Burrows LL. Forging new antibiotic combinations under iron-limiting conditions. Antimicrob Agents Chemother. 2020;64:e01909–19.
Singh PK. Iron sequestration by human lactoferrin stimulates P. aeruginosa surface motility and blocks biofilm formation. BioMetals. 2004;17:267–70.
Minandri F, Imperi F, Frangipani E, Bonchi C, Visaggio D, Facchini M, et al. Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect Immun. 2016;84:2324–35.
Iatsenko I, Marra A, Boquete J, Peña J, Lemaitre B. Iron sequestration by transferrin 1 mediates nutritional immunity in Drosophila melanogaster. Proc Natl Acad Sci USA. 2020;117:1–9.
Chan DCK, Burrows LL. Thiocillin and micrococcin exploit the ferrioxamine receptor of Pseudomonas aeruginosa for uptake. 2020. https://doi.org/10.1101/2020.04.23.057471.
Degiacomi G, Personne Y, Mondésert G, Ge X, Mandava CS, Hartkoorn RC, et al. Micrococcin P1—a bactericidal thiopeptide active against Mycobacterium tuberculosis. Tuberculosis. 2016;100:95–101.
Zheng Q, Wang Q, Wang S, Wu J, Gao Q, Liu W. Thiopeptide antibiotics exhibit a dual mode of action against intracellular pathogens by affecting both host and microbe. Chem Biol. 2015;22:1002–7.
Chang PT, Rao K, Longo LO, Lawton ES, Scherer G, Van, et al. Thiopeptide defense by an ant’s bacterial symbiont. J Nat Prod. 2020;83:725–9.
Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–14.
Claesen J, Spagnolo JB, Ramos SF, Kurita KL, Byrd AL, Aksenov AA, et al. Cutibacterium acnes antibiotic production shapes niche competition in the human skin microbiome. 2019. https://doi.org/10.1101/594010.
Acknowledgements
We thank Christy Groves for assistance with the figures. Work on thiopeptides in the Burrows lab is funded by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN-2016-06521) and the Ontario Research Fund (RE07-048). DCKC was funded by an NSERC Canadian Graduate Scholarship—Master’s award and holds an Ontario Graduate Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chan, D.C.K., Burrows, L.L. Thiopeptides: antibiotics with unique chemical structures and diverse biological activities. J Antibiot 74, 161–175 (2021). https://doi.org/10.1038/s41429-020-00387-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-020-00387-x