Abstract
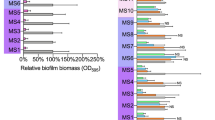
Interest has been rekindled in the old antibiotic fosfomycin, partly because of its ability to penetrate biofilm. Using a transcriptomic approach, we investigated the modifications induced by fosfomycin in sessile cells of a clinical Staphylococcus aureus isolated from a device-associated infection. Cells still able to form biofilm after 4 h of incubation in the presence of subinhibitory concentrations of fosfomycin and cells from 24-h-old biofilm later submitted to fosfomycin had 6.77% and 9.41%, respectively, of differentially expressed genes compared with their antibiotic-free control. Fosfomycin induced mostly downregulation of genes assigned to nucleotide, amino acid and carbohydrate transport, and metabolism. Adhesins and capsular biosynthesis proteins encoding genes were downregulated in fosfomycin-grown biofilm, whereas the murein hydrolase regulator lgrA and a d-lactate dehydrogenase-encoding gene were upregulated. In fosfomycin-treated biofilm, the expression of genes encoding adhesins, the cell wall biosynthesis protein ScdA, and to a lesser extent the fosfomycin target MurA was also decreased. Unattached cells surrounding fosfomycin-grown biofilm showed greater ability to form aggregates than their counterparts obtained without fosfomycin. Reducing their global metabolism and lowering cell wall turnover would allow some S. aureus cells to grow in biofilm despite fosfomycin stress while promoting hyperadherent phenotype in the vicinity of the fosfomycin-treated biofilm.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Del Pozo JL, Patel R. Infection associated with prosthetic joints. N Engl J Med. 2009;361:787–94.
Lebeaux D, Ghigo J-M, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78:510–43.
Cha J-O, Park Y-K, Lee YS, Chung GT. In vitro biofilm formation and bactericidal activities of methicillin-resistant Staphylococcus aureus clones prevalent in Korea. Diagn Microbiol Infect Dis. 2011;70:112–8.
Marquès C, Tasse J, Pracros A, Collin V, Franceschi C, Laurent F, et al. Effects of antibiotics on biofilm and unattached cells of a clinical Staphylococcus aureus isolate from bone and joint infection. J Med Microbiol. 2015;64:1021–6.
Siala W, Mingeot-Leclercq M-P, Tulkens PM, Hallin M, Denis O, Van Bambeke F. Comparison of the antibiotic activities of daptomycin, vancomycin, and the investigational fluoroquinolone delafloxacin against biofilms from Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother. 2014;58:6385–97.
Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, et al. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio. 2012;3:e00198–112.
Lázaro-Díez M, Remuzgo-Martínez S, Rodríguez-Mirones C, Acosta F, Icardo JM, Martínez-Martínez L, et al. Effects of subinhibitory concentrations of ceftaroline on methicillin-resistant Staphylococcus aureus (MRSA) biofilms. PLoS One. 2016;11:e0147569.
Ng M, Epstein SB, Callahan MT, Piotrowski BO, Simon GL, Roberts AD, et al. Induction of MRSA biofilm by low-dose β-Lactam Antibiotics: specificity, prevalence and dose-response effects. Dose Response. 2014;12:152–61.
Wang Q, Sun F-J, Liu Y, Xiong L-R, Xie L-L, Xia P-Y. Enhancement of biofilm formation by subinhibitory concentrations of macrolides in icaADBC-positive and -negative clinical isolates of Staphylococcus epidermidis. Antimicrob Agents Chemother. 2010;54:2707–11.
Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. In vivo pharmacokinetics/pharmacodynamics of colistin and imipenem in Pseudomonas aeruginosa biofilm infection. Antimicrob Agents Chemother. 2012;56:2683–90.
Tang H-J, Chen C-C, Cheng K-C, Toh H-S, Su B-A, Chiang S-R, et al. In vitro efficacy of fosfomycin-containing regimens against methicillin-resistant Staphylococcus aureus in biofilms. J Antimicrob Chemother. 2012;67:944–50.
Tang H-J, Chen C-C, Cheng K-C, Wu K-Y, Lin Y-C, Zhang C-C, et al. In vitro efficacies and resistance profiles of rifampin-based combination regimens for biofilm-embedded methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57:5717–20.
Barry AL, Brown SD. Antibacterial spectrum of fosfomycin trometamol. J Antimicrob Chemother. 1995;35:228–30.
Pérez DS, Tapia MO, Soraci AL. Fosfomycin: uses and potentialities in veterinary medicine. Open Vet J. 2014;4:26–43.
Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents. 2009;34:111–20.
Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. Fosfomycin. Clin Microbiol Rev. 2016;29:321–47.
Neuner EA, Sekeres J, Hall GS, van Duin D. Experience with fosfomycin for treatment of urinary tract infections due to multidrug-resistant organisms. Antimicrob Agents Chemother. 2012;56:5744–8.
Castañeda-García A, Blázquez J, Rodríguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics. 2013;2:217–36.
Marquès C, Franceschi C, Collin V, Laurent F, Chatellier S, Forestier C. Genome sequence of a clinical Staphylococcus aureus strain from a prosthetic joint infection. Genome Announc. 2016;4:2.
Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiol Read Engl. 2003;149:2719–32.
Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol. 2003;49:807–21.
Dengler V, Meier P, Heusser R, Berger-Bächi B, McCallum N. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol. 2011;11:16.
Shen F, Tang X, Wang Y, Yang Z, Shi X, Wang C, et al. Phenotype and expression profile analysis of Staphylococcus aureus biofilms and planktonic cells in response to licochalcone A. Appl Microbiol Biotechnol. 2015;99:359–73.
Groicher KH, Firek BA, Fujimoto DF, Bayles KW. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–801.
Petek M, Baebler S, Kuzman D, Rotter A, Podlesek Z, Gruden K, et al. Revealing fosfomycin primary effect on Staphylococcus aureus transcriptome: modulation of cell envelope biosynthesis and phosphoenolpyruvate induced starvation. BMC Microbiol. 2010;10:159.
Qin N, Tan X, Jiao Y, Liu L, Zhao W, Yang S, et al. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci Rep. 2014;4:5467.
Coldren FM, Palavecino EL, Levi-Polyachenko NH, Wagner WD, Smith TL, Smith BP, et al. Encapsulated Staphylococcus aureus strains vary in adhesiveness assessed by atomic force microscopy. J Biomed Mater Res A. 2009;89:402–10.
Dörries K, Schlueter R, Lalka M. Impact of antibiotics with various target sites on the metabolome of Staphylococcus aureus. Antimicrob Agents Chemother. 2014;58:7151–63.
Xu Y, Maltesen RG, Larsen LH, Schønheyder HC, Le VQ, Nielsen JL, et al. In vivo gene expression in a Staphylococcus aureus prosthetic joint infection characterized by RNA sequencing and metabolomics: a pilot study. BMC Microbiol. 2016;16:80.
Acknowledgements
CIFRE fellowship from bioMérieux and the Association Nationale de la Recherche et de la Technologie (ANRT) for CM is gratefully acknowledged. We thank Ophélie Chapeira, GenoScreen—Lille, for her help in RNA-seq data analysis. We would like to thank Anne Pracros for her technical help, Damien Balestrino and Cyril Guilhen for their helpful advice in statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Marquès, C., Collin, V., Franceschi, C. et al. Fosfomycin and Staphylococcus aureus: transcriptomic approach to assess effect on biofilm, and fate of unattached cells. J Antibiot 73, 91–100 (2020). https://doi.org/10.1038/s41429-019-0256-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-019-0256-y