Abstract
The β-lactams are the most widely used group of antibiotics in human health and agriculture, but this is under threat due to the persistent rise of pathogenic resistance. Several compounds, including tunicamycin (TUN), can enhance the antibacterial activity of the β-lactams to the extent of overcoming resistance, but the mammalian toxicity of TUN has precluded its use in this role. Selective hydrogenation of TUN produces modified compounds (TunR1 and TunR2), which retain the enhancement of β-lactams while having much lower mammalian toxicity. Here we show that TunR1 and TunR2 enhance the antibacterial activity of multiple β-lactam family members, including penems, cephems, and third-generation penicillins, to a similar extent as does the native TUN. Eleven of the β-lactams tested were enhanced from 2 to >256-fold against Bacillus subtilis, with comparable results against a penicillin G-resistant strain. The most significant enhancements were obtained with third-generation aminothiazolidyl cephems, including cefotaxime, ceftazidime, and cefquinome. These results support the potential of low toxicity tunicamycin analogs (TunR1 and TunR2) as clinically valid, synergistic enhancers for a broad group of β-lactam antibiotics.
Similar content being viewed by others
Introduction
Antibiotic-resistant bacteria are now responsible for two million infections per year in the US alone, of which more than 23,000 are lethal [1]. Overcoming infectious diseases caused by antibiotic-resistant bacterial pathogens is one of the major research priorities of the World Health Organization [2]. One approach to combating resistance is the use of drug combinations that exert a synergistic enhancement. Important examples of this are combination therapies of β-lactam antibiotics (penicillins, penems, and cephalosporins) with β-lactamase inhibitors, such as clavulanic acid, tazobactam, or sulbactam [3, 4]. Alternative strategies have focused on β-lactam enhancers that target bacterial cell wall biosynthesis [5,6,7]. These include synthetic compounds, such as tarocin A and tarocin B [7], and a Streptomyces-derived natural product, tunicamycin (TUN) [8, 9], which inhibit the assembly of cell wall teichoic acid (WTA) [7,8,9,10,11].
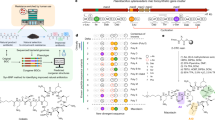
WTA, a cell surface carbohydrate polymer found in Gram-positive bacteria, is the target for several new antibacterial agents [4, 5]. The biosynthesis of WTA begins by transfer of GlcNAc-1-phosphate to a membrane-associated polyprenol, undecaprenyl phosphate, embedded in the bacterial surface. This transfer is catalyzed by the enzyme TagO (also called TarO), which is part of a large superfamily of phosphosugar transferases (PNPT) that also includes the first enzyme for N-linked protein glycosylation in eukaryotes, GPT, as well as MraY, an essential bacterial enzyme involved in peptidoglycan biosynthesis [12, 13]. TUN is a transition state analog inhibitor of the PNPT enzyme superfamily (Fig. 1), and as such it is a potent inhibitor of both eukaryotic protein N-glycosylation and the assembly of bacterial cell walls [14,15,16,17,18,19]. Thus although TUN is a potent antibacterial agent, it is also highly toxic to eukaryotic cells.
a Bacterial PNPT-catalyzed reaction of UDP-GlcNAc and undecaprenyl phosphate. b Schematic structure of tunicamycin, with arrows indicating the double bonds modified by hydrogenation to give TunR1 or TunR2. c Binding interaction mediated by π–π stacking between the uridyl group of tunicamycin and Phe2491 in the PNPT uridyl-binding pocket [23]. The nonplanarity of the 5,6-dihydrouridyl moiety of TunR2 reduces this binding interaction, resulting in lower mammalian toxicity [9]. 1Numbering based on hGPT. Where Tun/TunR2 is the tunicaminyl core
Structural and biochemical studies have shown that the eukaryotic (GPT, Alg7) and bacterial (MraY, WecA, TagO, etc.) PNPT family members are transmembrane proteins, which are respectively located in the endoplasmic reticulum (ER) or the (inner) bacterial membrane, with the TUN-binding site exposed at the lumen/cytoplasmic face [20,21,22,23,24,25,26]. Significantly, the tunicaminyl uracil group acts as a substrate analog of the UDP-HexNAc donor substrate of PNPT, and binds to a uridyl-binding pocket within the PNPT family ([9, 23, 26]; Fig. 1). The binding of TUN to PNPT is stabilized by a noncovalent π–π stacking interaction with a conserved phenylalanine residue (Phe) within the uridyl-binding pocket on the cytoplasmic side of the membrane (Fig. 1). A corresponding Phe is conserved in all PNPT family members, although sequences proximal to this differ between the eukaryotic and bacterial uridyl-binding sites [9, 23, 26]. There is also secondary binding between the GlcNAc moiety of TUN and the carbohydrate recognition domain on PNPT [26]. This is located on a cytoplasmic loop (loop E for hGPT or bacterial cytoloop 5) and directs recognition of the appropriate sugar nucleotide substrate, either UDP-GlcNAc or UDP-MurNAc-pentapeptide, respectively. Lastly, the isoprenyl substrate requirement for the eukaryotic PNPTs is highly specific for dolichol phosphates. These are generally longer chain prenols than the corresponding bacterial undecaprenyl phosphate, and also contain a saturated terminal α-isoprene unit [27,28,29]. The conjugated double bond in the N-acyl chain of TUN may mimic the unsaturated prenol phosphate substrate in the PNPT active site, partially determining the inhibitor binding to some degree [9, 26].
Based on these observations we have previously shown that chemically modifying the native TUN, either by selectively hydrogenating the N-acyl double bond (to give TunR1) or hydrogenating both the N-acyl and uridyl double bonds (giving TunR2) leads to compounds that are considerably less toxic to eukaryotic cells, but which retain their antibacterial activity [9]. Compared with native TUN, TunR1 and TunR2 are equally effective in enhancing the activity of three β-lactams, oxacillin, methicillin, and penicillin G, however, their inhibitory activity is significantly reduced in a Pichia-based protein N-glycosylation bioassay [9].
In this study, we first investigated the toxicity of the native and modified TUNs using live insect larvae as a model eukaryote. This was followed by studies of the enhancement of 12 β-lactam family members, including penems, cephems, and third-generation penicillins. In every case the nontoxic-modified TunR1 and TunR2 gave enhancements equivalent to or better than the toxic, unmodified TUN. Eleven of the lactams tested were enhanced from 2- to >256-fold against Bacillus subtilis, as a model Gram-positive bacterium, supported with comparable results against a penicillin G-resistant strain of B. subtilis. The penicillin biosynthetic precursor, aminopenicillanic acid (APA), which is generally considered to be non-antibacterial, had TUN-enhanced Minimal inhibitory concentrations (MIC) values in the low µg ml−1 range. Interestingly, large and selective MIC enhancements in excess of 128-fold were observed for three aminothiazolidyl-type cephems, while the non-thiazolidine cephalexin showed relatively little enhancement.
Results
Relative toxicity of tunicamycin, TunR1, and TunR2 against an insect larvae model
We evaluated the relative toxicity of the TUN-derived compounds against a live insect larvae as a simple animal model. TunR1 and TunR2 have been previously shown to have significantly reduced toxicity against eukaryotic cell lines (Chinese hamster ovary CHO and human MDA-MB-231) compared with native TUN [9] but this has not been previously assessed for a live animal model. To evaluate this, first instar fall armyworm (Spodoptera frugiperda) larvae were grown on insect diet disks containing the various TUN compounds (15 µg per disk). The larvae growth was visually assessed and weighed after 3 days (Fig. 2). The native TUN visually inhibited the growth of the larvae, and reduced the mean weight/insect by about 75% (from 0.82 to 0.18 mg). Both TunR1 and especially TunR2 were considerably less toxic towards the larvae. The TunR1 reduced larvae growth by ~50%, whereas the TunR2 is essentially nontoxic at this concentration (Fig. 2). Further studies in higher animal models are needed to substantiate the claim that R1 and R2 are less toxic than Tun.
Antibacterial enhancement of penicillin and penems by TUNs
A microtiter-based penicillin enhancement bioassay [9] was used to assess the antibacterial activity of 11 commercially available β-lactam drugs (plus d-cycloserine) in combinations with native TUN, and two modified tunicaminyl compounds, TunR1 and TunR2. Four second-generation penicillins (amoxicillin, ampicillin, carbenicillin, and piperacillin), four cephems (cefotaxime, cefquinome, ceftazidime, and cephalexin) including one exclusively used in veterinary medicine (cefquinome), a carbapenem (imipenem), and the penicillin core APA, plus a penicillin/lactamase inhibitor combination drug (augmentin) were evaluated in the bioassay. d-cycloserine was included as a nonlactam antibiotic control that also targets peptidoglycan biosynthesis. MIC (Table 1) and minimal stationary inhibitory concentrations (MStC, Table S1) were measured from dilution series of these antibiotics using a resazurin-based live/dead stain. These were assessed in triplicate as either dead (blue) or stationary (pink) and were scored visually by four individuals. The MIC and MStC were assessed with respect to positives (Supplementary data Fig S3). The broth dilution MICs for TUN, TunR1, or TunR2 alone for B. subtilis are 0.15, 0.3, or 0.3 μg ml−1, respectively, whereas for Saccharomyces the respective MICs are 0.3, 2.5, or 410 μg ml−1 [9].
MICs for the penicillin group and imipenem against B. subtilis MW10 ranged from 0.125 to 0.5 µg ml−1, with relatively modest enhancement (less than eightfold) in the presence of the TUNs. The largest enhancements were 4–16-fold and 8–16-fold for imipenem and piperacillin, respectively, and are small compared with the 64-fold enhancements previously observed with oxacillin [8, 9]. These combinations were also tested on a penicillin G-resistant strain of MW10 (designated PGr). The imipenem enhancements are similar (eightfold) on the resistant PGr strain, but no enhancement was observed for piperacillin. (Table 1). The MStC were also similar for these combinations, although the enhancements were greater, and typically in the range from 16- to >64-fold. The most dramatic effect observed was with imipenem, for which the MStC was decreased from 0.0625 µg ml−1 to less than 0.001 µg ml−1, a >64-fold enhancement (Fig. 3a). Similar enhancement of the MStC occurred with both TunR1 or TunR2, and are comparable to those observed with the native TUN (Supplementary data Table S1).
Broth dilution enhancement assays. Imipenem (a) or 6-aminopenicillanic acid (APA) (b) on B. subtilis MW10 and the penicillin G-resistant mutant PGr. Vertical rows contain the β-lactam dilution gradient (imipenem, 0.001–0.125 µg ml−1; APA, (0.39–50.0 µg ml−1). The enhancers used are tunicamycin (rows 4–6), TunR1 (rows 7–9), or TunR2 (rows 10–12). MICs are shown by black bars. DMSO dimethylsulfoxide, negative control (rows 1–3)
The microtiter-based assay results described above were confirmed using an agar diffusion bioassay. Bacillus MW10 on TUN-containing agar (2 µg/20 ml or 5 µg/20 ml) was over-spotted with various β-lactams (1 µg of each) and assessed visually with resazurin after 12 h (Supplementary Fig. S2). Zones of clearance were evident at high (5 µg/20 ml) and low (2 µg/20 ml) concentrations of the TUNs (Fig. S2A, B). However, the pink coloration of the resazurin overlay revealed that bacteria were present at the lower concentration (Fig. S2C), as agar-embedded spores. At the high concentration the resazurin-stained zones are blue, indicating inhibition of growth by the various penicillin/TUN combinations (Fig. S2D).
Antibacterial enhancement of 6-aminopenicillanic acid
APA is the structural core of the penicillin molecule, composed of the fused five-member thiazolidine and four-membered lactam rings, and is typically produced by enzymatic or chemical hydrolysis of the benzyl group of penicillin G [30, 31]. It is used commercially as the starting material for the preparation of numerous semisynthetic penicillins, but is generally considered to have poor antibiotic activity itself [32]. In agreement with this, APA was found to have a high MIC against B. subtilis MW10 (12.5 µg ml−1), and especially so against PGr (50.0 µg ml−1) (Fig. 3b and Table 1). This was also observed for the MStC with 25 µg ml−1 for the PGr, and no observable “pink” stationary wells (Supplementary Table S1). However, in presence of the various TUN enhancers the MICs were notably reduced into the low µg/ml range, and even less so for the MStC (Supplementary Tables S1 and S2). The APA/TunR1 combination displayed a MIC value of 1.56 µg ml−1 against MW10, an eightfold enhancement compared with APA alone. Against PGr, APA/TUN and APA/TunR2 gave the most potent enhancements with improved MICs of 6.25 µg ml−1 and typical MStC values of 0.4 µg ml−1. Hence, the penicillin biochemical precursor APA is enhanced in the presence of the modified TUNs to give useful antibacterial activity in the low micromolar range.
Antibacterial enhancement of cephalosporins (Cephems) by TUNs
The ability of TUNs to enhance cephalosporins has not been tested previously and we therefore evaluated this for four commonly used cephems: cephalexin, cefotaxime, cefquinome, and ceftazidime (Tables 1 and S1). The relative enhancement of MICs and MStCs were assessed for sub-MIC concentrations of native TUN, TunR1, and TunR2. The dramatic enhancement of three of the cephems (Cef, Ceq, and Cez) needed a modification of the bioassay, requiring two microtiter plates to cover the entire dynamic range (examples shown in Supplementary Fig. S3). Hence, MIC enhancements were observed in the >128–256-fold range for these antibiotics, and are far greater that the twofold enhancement observed for the first-generation cephalexin (Fig. S3B).
Similar results were obtained against the penicillin G-resistant PGr strain, with the third-generation cephems having the largest MIC enhancements (Table 1). The minimal stationary concentrations (MStC) for the β-lactams were also significantly enhanced by the modified TUNs (Table S1). This was most noticeable for imipenem, piperacillin, and carbenicillin, which were >32-fold enhanced by TUN, TunR1, and TunR2. The MStC for APA was enhanced 64-fold by TunR1 or TunR2 against the wild type MW10, and by >128-fold by the native TUN. Also unexpected was the large MStC enhancement (>256-fold in combination with TunR2) seen for the non-β-lactam drug, d-cycloserine.
Checkerboard cross-activity bioassays
The most significantly enhanced cephems (cefotaxime and cefazidime) were further evaluated using checkerboard cross-activity bioassays with TunR1, TunR2, and the native TUN (Supplementary Figs. S4 and S5, and exemplified in Fig. 4). The minimum inhibitory concentrations (MIC90) of the individual compounds, the lowest MIC90 achieved in the various combinations of TUN derivatives with cephems, and the fractional inhibitory concentration indices (FICI) are summarized in Fig. 4. Interactions obtained for the native TUN or its derivatives with cefotaxime or ceftazidime resulted in synergism with FICI < 0.5 (Fig. 4). Combinations of TunR1 with cefotaxime exhibited the highest synergistic interaction with FICI of 0.14. TunR2 exhibited a similar level of synergy with both cefotaxime and ceftazidime, 0.185 and 0.187, respectively. Hence, the TUN derivatives showed a strong synergy with the third-generation cephems, cefotaxime, or ceftazidime.
The checkerboard assay of ceftazidime (0–10.0 µg ml−1) verses TunR2 (0–0.5 µg ml−1). The adjacent table shows FICI values for cefotaxime (Cefo) and ceftazidime (Ceftaz) in combination with TunR2. 1Fractional inhibitory concentration index (FICI) are calculated from FICI = (MIC drug A in combination/MIC drug A alone) + (MIC drug B in combination/MIC drug B alone). Drug synergy is defined as an FICI ≤ 0.5, additivity/indifference is defined as an FICI > 0.5–4, and antagonism is defined as an FICI > 4
Discussion
The β-lactam class of antibiotics are often divided into four groups: penicillins (penams), cephalosporins (cephems), monobactams and carbapenems (penems), and further subdivided by generations. The availability of 6-APA, the penicillin structural core, facilitated the development of many semisynthetic β-lactams, and this is now the most prevalent use of bulk penicillin production [31]. The cephems were introduced because of their effectiveness against β-lactamase-derived resistance, and chemical modifications of the 7-aminocephalosporanic acid core has resulted in several “generations” of new cephems [33]. Cephalexin (Keflex®) is a first-generation aminobenzyl-type cephem, analogous to the ampicillin-type penicillins, while cefotaxime and ceftazidime are third-generation aminothiazolidine-type lactams. The C7-aminothiazolidyl/oxyimino cephem family was introduced for improved activity against certain β-lactam-resistant Gram-negative pathogens [34], and the syn-imino/aminothizolidyl substituent also has a protective effect against most CTX-M β-lactamases [35]. Cefquinome (Cobactan®) is often considered a fourth-generation cephem and is currently used exclusively in veterinary medicine [34].
There are several compounds that synergistically enhance the activity of the β-lactam antibiotics, but to date the majority of these are β-lactamase inhibitors [35]. Benzimidazole analogs have been developed as inhibitors of WTA biosynthesis, and have a potent synergistic effect in combination with imipenems [36]. Similarly, inhibiting WTA biosynthesis with short, branched-chain polyethyleimines enhances β-lactam efficacy against MRSA, but not against nonresistant Bacillus [37]. The TUNs are the most potent enhancers of β-lactam antibiotics found to date, but their clinical use is limited by the eukaryotic toxicity [7,8,9]. The blocking of protein N-glycosylation by TUN results in the accumulation of misfolded proteins in the ER and causes ER stress, leading to the lethal unfolded protein response (UPR). Native TUN caused UPR in the cerebral cortex, hippocampus, and cerebellum of mice [38]. TUN and the related corynetoxins often cause lethal annual ryegrass toxicity in grazing livestock. Sheep are especially sensitive to these toxins with a lethal dose of about 35 µg kg−1 bodyweight for pure TUN given by subcutaneous injection and 3–5 mg kg−1 for corynetoxin administered orally [39]. We have previously reported that selectively hydrogenated TUNs (TunR1 and TunR2) are considerably less toxic to yeast or mammalian cell lines than are the native TUNs but retain the β-lactam enhancement activity [9]. TunR2, in particular, is essentially nontoxic (MIC > 100 μg ml−1) against eukaryotic cells [9]. By contrast, native TUNs block growth with typical MIC 0.2–0.5 μg ml−1 against these cell types. In the present paper we have determined that TunR2 is nontoxic to live insect larvae, and may overcome the toxicity issues normally incurred with natural TUN.
In bacteria, TUN targets PNPP enzymes in the TagO and MraY enzyme families that catalyze the early steps of WTA biosynthesis and peptidoglycan assembly, respectively. The β-lactam group of antibiotics also block peptidoglycan assembly, but at a later step involving cross-linking by penicillin binding proteins (PBPs). Most bacteria have several forms of PBPs (B. subtilis has at least seven [40]), many of which have diverse roles associated with cell division, growth, and assembly of the division septum [41]. All PBPs carry a catalytic triad in which the active site serine makes a covalent adduct with the β-lactam antibiotics. However, several of the β-lactams are known to have differential activity against different PBPs, even within the same cell [41]. PBP2B is the lethal target for β-lactams in Bacillus and has a high affinity for methicillin and ampicillin [40]. Cephamycin C has lower affinity for PBP2B but high affinity for PBP1, and has a weak anti-Bacillus activity. Mutations resulting in the loss of PBP3 or PBP2A makes B. subtilis generally more sensitive to β-lactams [41]. However, mutants lacking PBP3 showed increased sensitivity to oxacillin and cephalexin compared with wild type, but not to penicillin G, suggesting that different PBPs are targeted by these drugs [41]. Indeed, combinations of β-lactams that target different PBPs are also known to synergistically enhance each other [42].
The correct cellular location of the respective B. subtilis PBPs during cell division is critical, and driven, at least in part, by the localization of lipid II, the precursor substrate for the peptidoglycan cross-linking reaction [43]. TUN inhibition of MraY may alter the cellular lipid II pool by blocking production of its immediate metabolic precursor, lipid I, thereby resulting in inappropriate localization of key PBPs. However, this altered lipid I/lipid II localization is unlikely to be a major mechanism for the enhancement of the β-lactam drugs against these key PBPs. This is because TUN inhibition of MraY is relatively poor compared with the inhibition of TagO [21, 26], and because the TUN enhancement was not observed for Gram-negative bacteria that lack WTA entirely. A more likely model is that TUN inhibition of TagO results in depleted WTA synthesis. Newly synthesized WTA recruits PBP4 to the site of cell division, where it completes the cross-linking of peptidoglycan formed by PBP2/PBP2a. The mislocation of these key PBPs by the depletion of WTA may result in the additional sensitivity of the bacterial cells to β-lactams.
In the present paper we have shown that less-toxic TUN analogs, TunR1 and TunR2, effectively enhance a large range of β-lactam antibiotics against wild type and resistant B. subtilis. In particular, we found a dramatic enhancement for aminothiazolidine-type cephems, cefotaxime, ceftazidime, cefquinome, in the order of 125–250-fold. Low FICI values were observed for cephem-TUN combinations, and are highly indicative of potent synergistic enhancement. Cefquinome is used extensively in veterinary medicine to treat bovine respiratory disease, one of the most commercially important diseases in cattle. Typically, an intravenous dose of 10 mg kg−1 is used in cattle and pigs, hence requiring gram quantities for treatment of calves [32]. For pigs, cefquinome is used to treat respiratory infections and against meningitis caused by Streptococcus suis, which is endemic in all countries with an extensive pig industry [34]. A combination drug of TunR2-enhanced cefquinome resulting in a 125-fold improvement of antibacterial activity could significantly lower costs to the farming industry and, our result suggest, may provide sufficient potency to overcome certain forms of β-lactam resistance.
Materials and methods
Materials
TUN was purchased from Alfa Aesar, Ward Hill, MA. TunR1 and TunR2 were prepared by selective catalytic hydrogenation of TUN as described previously [9], either using 5 wt% Pd/C (Calsicat, Erie, PA) or 5 wt% Rh/Al2O3 (BASF Engelhard, Iselin, NJ), as the respective catalysts. All other chemicals, including culture media, were obtained from Sigma-Aldrich Inc., St Louis, MO, USA.
Analytical methods
Matrix-assisted laser desorption/ionization time-of-flight mass spectra were recorded on a Bruker Daltonics Microflex instrument (Bruker Daltonics, Billerica, MA, USA) as described [44]. NMR spectra were obtained on a Bruker Avance III spectrometer (Bruker BioSpin) operating at 500.11 MHz. Samples were dissolved in deuterated methanol. HPLC analyses were undertaken on a Finnigan Surveyor instrument (ThermoFisher Scientific, West Palm Beach, FL, USA) using a C-18 Spheri-5 ODS column (250 mm, 5 mm particle size) eluted with aqueous acetonitrile (45 to 100% v/v; 15 min, 1 ml min−1) [45].
Insect larvae toxicity bioassay
Bioassays were performed essentially as described previously [46]. Briefly, 30 µl of a 0.5 µg µl−1 methanol solution of each test compound was applied to a 15 mg freeze-dried insect diet disk, and the disks were placed in a fume hood for 1 h to evaporate the methanol. Then 30 µl of sterile water was added to the disk, which was allowed to equilibrate for 30 min in the assay dish. Ten first instar fall armyworm (S. frugiperda) larvae were added to each Petri dish containing the disk. Treatments were assayed in duplicate. The larvae were allowed to feed on the treated diet disk for 3 days, and were then weighed using an analytical balance. Data were analyzed for statistically significant differences using analysis of variance program Proc GLM, SAS version 8.0.
β-Lactam antibacterial enhancement assays
MIC enhancements for the various β-lactam antibiotics were measured in the presence of a sublethal quantity of the TUN samples under test using a microtiter plate-based bioassay [9]. The assays were done in sterile 96-well round-bottom microtiter plates (Cellstar, Greiner Bio-one, Munroe, NC). Aqueous stocks (1 mg ml−1) of the β-lactam under test were prepared in 10 ml volumetric flasks, and stored at 4 °C. These were diluted to the appropriate concentrations by diluting to 6 ml with sterile culture medium, immediately before the assay. The β-lactam dilutions (200 µl) were added to the top wells (Row A) of the plates, and 100 µl of sterile tryptic soy broth medium to the remaining wells. Dilution gradients were constructed by serially dilution of 100 µl from the Row A wells into the lower rows. The microbial inoculums were prepared in phosphate-buffered saline (10 ml, 0.5 McFarland) using a turbidometer. This was diluted 125 µl to 12.5 ml (1–100 v/v) with sterile media. Resazurin stock solution (25 µl of 10 mg/200 µl in 50:50 MeOH:water) was added, and then mixed to give a dark blue color. The inoculum was transferred to a trough, and 100 µl dispensed into the β-lactam-containing microtiter plate wells using a multichannel pipet. The plates were incubated at 28 °C for 24 h. Lids were placed on the plates to ensure they do not dry out. MICs and MStCs were assessed visually from the color change of the resazurin live/dead stain.
For the enhancement experiments the microtiter plates were divided into four sections, each comprised of three columns (1–3, 4–6, 7–9, and 10–12). Native TUN (0.2 μg ml−1, final concentration) was added to the wells in columns 4–6, TunR1 (0.4 μg ml−1) in 7–9, or TunR2 (0.4 μg ml−1) in 10–12. The TUN-free control wells (columns 1–3) received 10 μL of DMSO, the stock solution solvent.
Checkerboard drug susceptibility testing
MIC testing was carried out by broth microdilution using the AlamarBlue (AB, BioRad) assay [47]. For pairwise combination (checkerboard) assays, a two-dimensional array of serial dilutions of the compounds was prepared in 96-well plates, as previously described [47, 48]. Relative MIC90 concentrations in wells representing various ratios of the two compounds were used for calculations to determine whether paired combinations exert inhibitory effects that are more than the sum of their effects alone (synergy). Cell viability was then determined and the FICI values that correspond to either compound (A or B) were MIC90 of A (combination)/MIC90 of A (singly) + MIC90 of B (combination)/MIC90 of B (singly), as defined by the Odds [49].
References
CDC Centers for Disease Control and Prevention. https://www.cdc.gov/drugresistance/
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther 2015;40:277–83.
Bush K. Game changers: new β-lactamase inhibitor combinations targeting antibiotic resistance in Gram-negative bacteria. ACS Infect Dis. 2018;4:84–7.
Pasquina LW, Santa Maria JP, Walker S. Teichoic acid biosynthesis as an antibiotic target. Curr Opin Microbiol. 2013;16:531–7.
Sewell EW, Brown ED. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot. 2014;67:43–51.
Labroli MA, Caldwell JP, Yang C, Lee SH, Wang H, Koseoglu S, et al. Discovery of potent wall teichoic acid early stage inhibitors. Discovery of potent wall teichoic acid early stage inhibitors. Bioorg Med Chem Lett. 2016;26:3999–4002.
Lee SH, Wang H, Labroli M, Koseoglu S, Zuck P, Mayhood T, et al. TarO-specific inhibitors of wall teichoic acid biosynthesis restore β-lactam efficacy against methicillin-resistant staphylococci. Sci Transl Med. 2016;8:329ra32.
Campbell J, Singh AK, Santa Maria JP, Kim Y, Brown S, Swoboda JG, et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol. 2011;6:106–16.
Price NPJ, Hartman TM, Li J, Velpula KK, Naumann TA, Guda MR, et al. Modified tunicamycins with reduced eukaryotic toxicity that enhance the antibacterial activity of β-lactams. J Antibiot. 2017;70:1070–7.
Hancock IC, Wiseman G, Baddiley J. Biosynthesis of the unit that links teichoic acid to the bacterial wall: Inhibition by tunicamycin. FEBS Letts. 1976;69:75–80.
Kern J, Ryan C, Faull K, Schneewind O. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J Mol Biol. 2010;401:757–75.
Price NP, Momany FA. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15:29R–42R.
Al-Dabbagh B, Olatunji S, Crouvoisier M, El Ghachi M, Blanot D, Mengin-Lecreulx D, et al. Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie. 2016;127:249–57.
Tamura G. Tunicamycins. Tokyo, Japan: Japan Scientific Press; 1982.
Price NPJ, Tsvetanova B. Biosynthesis of the tunicamycins: a review. J Antibiot. 2007;60:485–91.
Heifetz A, Keenan RW, Elbein AD. Mechanism of action of tunicamycin on the UDP-GlcNAc: dolichyl-phosphate GlcNAc-1-phosphate transferase. Biochemistry. 1979;18:2186–92.
Mclachlan KR, Krag SS. Substrate specificity of N-acetylglucosamine 1-phosphate transferase activity in chinese hamster ovary cells. Glycobiology. 1992;2:313–9.
Eckardt K. Tunicamycins, streptovirudins, and corynetoxins, a special subclass of nucleoside antibiotics. J Nat Prod. 1983;46:544–50.
Price NPJ, Labeda DP, Naumann TA, Vermillion KE, Bowman MJ, Berhow MA, et al. Quinovosamycins: New tunicamycin-type antibiotics in which the α, β-1′,11′-linked N-acetylglucosamine residue is replaced by N-acetylquinovosamine. J Antibiot. 2016;69:637–46.
Hering J, Dunevall E, Ek M, Brändén G. Structural basis for selective inhibition of antibacterial target MraY, a membrane-bound enzyme involved in peptidoglycan synthesis. Drug Discov Today. 2018;23:1426–35.
Brandish PE, Kimura K-I, Inukai M, Southgate R, Lonsdale JT, Bugg TDH. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob Agents Chemother. 1996;40:1640–4.
Xu L, Appell M, Kennedy S, Momany FA, Price NPJ. Conformational analysis of chirally deuterated tunicamycin as an active site probe of UDP-N-acetylhexosamine:polyprenol-P N-acetylhexosamine-1-P translocases. Biochemistry. 2004;43:13248–55.
Hakulinen JK, Hering J, Brändén G, Chen H, Snijder A, Ek M, et al. MraY-antibiotic complex reveals details of tunicamycin mode of action. Nat Chem Biol. 2017;13:265–7.
Chung BC, Zhao J, Gillespie RA, Kwon D-Y, Guan Z, Hong J, et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–6.
Chung BC, Mashalidis EH, Tanino T, Kim M, Matsuda A, Hong J, et al. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature. 2016;533:557–60.
Yoo J, Mashalidis EH, Kuk ACY, Ichikawa S, Lee S-Y. GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation. Nat Struct Mol Biol 2018;25:217–24.
Lukose V, Walvoort MTC, Imperiali B. Bacterial phosphoglycosyl transferases: initiators of glycan biosynthesis at the membrane interface. Glycobiology. 2017;27:820–33.
Jones MB, Rosenberg JN, Betenbaugh MJ, Krag SS. Structure and synthesis of polyisoprenoids used in N-glycosylation across the three domains of life. Biochim Biophys Acta. 2009;1790:485–94.
Cantagrel V, Lefeber DJ, Ng BG, Guan Z, Silhavy JL, Bielas SL, et al. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell. 2010;142:203–17.
Rolinson GN. The influence of 6-aminopenicillanic acid on antibiotic development. J Antimicrob Chemother. 1988;1988:5–14.
Elander RP. Industrial production of beta-lactam antibiotics. Appl Microbiol Biotechnol. 2003;61:385–92.
Petri Jr WA. Penicillins, cephalosporins, and other beta-lactam antibiotics. In: Brunton L Chabner, B Knollman, editors. Goodman and Gilman’s the pharmacological basis of therapeutics, (12th ed.). New York: McGraw Hill Professional; 2011. p. 1477–504.
Poirel L, Naas T, Le Thomas I, Amal K, Bingen E, Nordmann P. CTX-M-type extended-spectrum β-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob Agents Chemother. 2001;45:3355–61.
Limbert M, Isert D, Klesel N, Markus A, Seeger K, Seibert G, et al. Antibacterial activities in vitro and in vivo and pharmacokinetics of cefquinome (HR llyV), a new broad-spectrum cephalosporin. Antimicrob Agents Chemother. 1991;35:14–9.
Tehrani KHME, Martin NI. β-lactam/β-lactamase inhibitor combinations: an update. Medchemcomm. 2018;17:1439–56.
Yang S-W, Pan J, Yang C, Labroli M, Pan W, Caldwell J, et al. Benzimidazole analogs as WTA biosynthesis inhibitors targeting methicillin resistant Staphylococcus aureus. Bioorg Med Chem Lett. 2016;26:4743–7.
Foxley MA, Wright SN, Lam AK, Friedline AW, Strange SJ, Xiao MT, et al. Targeting wall teichoic acid in situ with branched polyethylenimine potentiates β-lactam efficacy against MRSA. ACS Med Chem Lett. 2018;8:1083–8.
Wang H, Wang X, Ke ZJ, Comer AL, Xu M, Frank JA, et al. Tunicamycin-induced unfolded protein response in the developing mouse brain. Toxicol Appl Pharmacol. 2015;283:157–67.
Jago MV, Culvenor CC. Tunicamycin and corynetoxin poisoning in sheep. Aust Vet J. 1987;64:232–5.
Horikawa S, Ogawara H. Penicillin-binding proteins in Bacillus subtilis. The effects on penicillin-binding proteins and the antibacterial activities of beta-lactams. J Antibiot. 1980;33:614–9.
Sassine J, Xu M, Sidiq KR, Emmins R, Errington J, Daniel RA. Functional redundancy of division specific penicillin-binding proteins in Bacillus subtilis. Mol Microbiol. 2017;106:304–18.
Foster TJ. Can β-lactam antibiotics be resurrected to combat MRSA? Trends Microbiol. 2019;27:26–38.
Lages MC, Beilharz K, Morales Angeles D, Veening JW, Scheffers DJ. The localization of key Bacillus subtilis penicillin binding proteins during cell growth is determined by substrate availability. Environ Microbiol. 2013;15:3272–81.
Price NPJ, Jackson MA, Vermillion KE, Li J, Yu B. Selective catalytic hydrogenation of the N-acyl and uridyl double bonds in the tunicamycin family of protein N-glycosylation inhibitors. J Antibiot. 2017;70:1122–8.
Tsvetanova BC, Price NPJ. Liquid chromatography-electrospray mass spectrometry of tunicamycin-type antibiotics. Anal Biochem. 2001;289:147–56.
Dowd PF, Berhow MA, Johnson ET. Differential activity of multiple saponins against omnivorous insects with varying feeding preferences. J Chem Ecol. 2011;37:443–9.
Singh V, Brecik M, Mukherjee R, Evans JC, Svetlikova Z, Blasko J, et al. The complex mechanism of antimycobacterial action of 5-fluorouracil. Chem Biol. 2015;22:63–75.
Singh V, Donini S, Pacitto A, Sala C, Hartkoorn RC, Dhar N, et al. The inosine monophosphate dehydrogenase, GuaB2, is a vulnerable new bactericidal drug target for tuberculosis. ACS Infect Dis. 2017;3:5–17.
Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52:1.
Acknowledgements
We thank David Lee for technical assistance and Dr Christopher Skory for pre-review of the manuscript. VS acknowledges the South African Medical Research Council (SAMRC) for funding. Mention of any trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Contributions
NPJP, MAJ, PFD and VS designed research; NPJP, MAJ, VS, PFD, TMH and JAB performed research; NPJP, MAJ, and VS analyzed the data; and NPJP, MAJ, VS, PFD and TMH wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Price, N.P.J., Jackson, M.A., Singh, V. et al. Synergistic enhancement of beta-lactam antibiotics by modified tunicamycin analogs TunR1 and TunR2. J Antibiot 72, 807–815 (2019). https://doi.org/10.1038/s41429-019-0220-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-019-0220-x