Abstract
Two new diketopiperazines (1, 2), one new polyprenol (3), together with 19 known compounds (4–22) were obtained from the EtOAc extract of Bionectria sp. Y1085, an endophytic fungus isolated from the plant Huperzia serrata. Their structures were elucidated by extensive NMR and MS analysis. Bionectin D (1) is a rare diketopiperazine with a single methylthio substitution at the α-carbon of cyclized amino acid residue. The antibacterial activity of compounds was assayed against Escherichia coli, Staphylococcus aureus, and Salmonella typhimurium ATCC 6539, and some metabolites (1, 2, 10, 11, and 14) exhibited evident antibacterial activity.
Similar content being viewed by others
Introduction
Fungi are biosynthetically talented organisms which can produce various amazing molecules with intricate architectures and promising bioactivities, and their fascinating structures and potential in drug development have gained increasing attention from scientists in natural product chemistry [1]. Endophytic fungi widely reside in all kinds of plants, and play a variety of roles ranging from symbiotic to pathogenic [2]. The struggles for livelihood, unique biotope provided by host and continual metabolic interactions with host resulted in an accumulation of unique substances produced by endophytic fungi to protect the host organism [3, 4]. Those bioactive metabolites are candidates for treating newly developing diseases, especially in the area of infection, cancer, and angiocardiopathy [5].
Previous investigations have shown that the metabolites produced by endophytic fungi are affected by host plant to some extent, thus the exploration of chemical constituents of endophytic fungi from medicinal plants is an effective way for drug discovery [6]. Huperzia serrata, a Chinese folk medicinal herb, is well-known source of huperzine A, a potent, highly specific, and reversible inhibitor of acetylcholinesterase [7]. As a part of our ongoing search for new active compounds from endophytic fungi in H. serrata [8, 9], a chemical investigation on the strain Bionectria sp. Y1085 was performed. As a result, two new diketopiperazines (1, 2), one new polyprenol (3), together with 19 previously reported compounds (4–22) were successfully obtained and identified. Among them, bionectin D (1) is a rare dioxopiperazine with a single methylthio substitution at the α-carbon of a cyclized amino acid residue. Moreover, the antibacterial activity of compounds was assayed against Escherichia coli, Staphylococcus aureus, and Salmonella typhimurium ATCC 6539, and some metabolites (1, 2, 10, 11, and 14) exhibited antibacterial activity. Herein, we describe the isolation and structural elucidation and the antibacterial activity of the isolated compounds.
Results and discussion
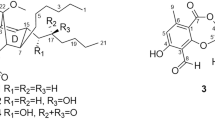
Bionectin D (1) was obtained as a colorless amorphous powder. Its molecular formula C16H19N3O3S was determined based on the HR-EI-MS signal at m/z 333.1148 [M]+, requiring nine degrees of unsaturation. A close inspection of the 1H, 13C NMR, and DEPT (Table 1) data of 1, and in combination with the HSQC spectrum suggested the presence of a 1,2-disubstituted benzene ring [δH 7.65 (1H, d, J = 8.0 Hz), δC 120.1; δH 6.98 (1H, t, J = 8.0 Hz), δC 120.2; δH 7.05 (1H, t, J = 8.0 Hz), δC 122.4; and δH 7.29 (1H, d, J = 8.0 Hz), δC 112.1], one olefinic methine (δH 7.10, 1H, s), one methylene (δH 3.65, 3.14, 1H each, ABq, J = 14.2 Hz), one singlet methyl (δH 2.23, s), one doublet methyl (δH 1.05, d, J = 6.7 Hz), two carbonyls (δC 168.8, 169.6), two sp3 methine (δC 61.2 and 68.3), three sp2 quaternary (δC 108.7, 128.8, and 137.6), and one sp3 quaternary (δC 70.3). A combination of the 1H and 13C NMR date with the degree of saturation imply that 1 possesses a diketopiperazine skeleton which derivatives with an indolylmethyl side chain, commonly isolated from the fungi of the genus Bionectria [10]. Detailed comparison of the NMR data with those of glioperazine C [10] and glioperazine [11] revealed that 1 is an indolylmethyl diketopiperazine, resulting from a cyclization of a threonine and a tryptophan with a methylthio substitution at C-6 (δC 70.3) (Fig. 1). This deduction was confirmed by the COSY and the HMBC correlations (Fig. 2): the 1H−1H COSY spectrum of compound 1 revealed two fragments as shown in Fig. 2 by the clear correlations of H-3/H-15/H-16 and H-11/H-12/H-13/H-14, respectively; the HMBC correlations (Fig. 2) showed a threonine and a tryptophan which contained 1,2-disubstituted benzene ring and replaced by a methylthio substitution at C-6: correlations from S-Me protons (δH 2.23) to C-6 (δC 70.3) and from H-3 (δH 2.23) to C-1 (δC 169.6), C-4 (δC 168.8), and C-15 (δC 68.3). The relative configuration of 1 was elucidated by comparing the chemical shifts of H-3 and H-16 with those reported data [10, 12] as well as by the analysis of ROESY correlations (Fig. 3). Previous studies have shown that the chemical shifts of the protons that located on the same side of diketopiperazine ring with an aromatic ring suffer an upfield shift due to the anisotrophic effect of the aromatic ring [10, 12]. For 1, the chemical shift of H-3 was 2.86 ppm (Table 1), which suggesting that the indole ring and H-3 are cis and assigned as α-oriented, the same with glioperazine C [10,11,12]. Meanwhile, the observed key ROESY cross-peaks of S-Me/H-16 indicated the β-orientation of S-methyl and 1-hydroxyethyl side chain (Fig. 3). Thus, the structure of 1 was determined as shown, and it is a rare diketopiperazine metabolite with a single methylthio substitution at the α-carbon of cyclized amino acid residue.
Bionectin E (2) was isolated as a white powder with positive optical rotation ([α]\({\,\!}_{\mathrm{D}}^{24}\) + 417.4, c 0.14, MeOH). Its HR-ESI-MS showed a [M+Na]+ ion peak at m/z 501.1024, corresponding to the molecular formula of C24H22N4O3S2, which represented 16 degrees of unsaturation. The observed UV maxima at 203, 218, and 281 nm were characteristic absorptions of monomeric epidithiodioxopiperazines with an indole ring [13]; furthermore, the 1H and 13C NMR data of 2 (Table 1) showed the existence of many same functionalities appear in bionectin B, including 1,2-disubstituted benzene ring, two sp3 methines, one methylamino group, two amide carbonyls, as well as same numbers of sp2 and sp3 quaternary carbons, which suggested that the skeleton of 2 was same with the skeleton of bionectin B (8) [14]. The major difference was that the non-oxygenated methylene (δC 24.7, C-12) in 2 replaced the hydroxylated methine (δC 68.1, d) at same position of bionectin B (8) [14]. This deduction was confirmed by the observed COSY correlation from H-12 to H-13 and HMBC cross-peaks from H-12 to C-3 (δC 77.4), C-4 (δC 161.8) (Fig. 2). The observed ROESY correlations (Fig. 3) from H-10/H-11; H-5a/H-2′, H-4′ as well as the almost identical optical rotation values (bionectin B, [α]D, + 493, c 0.2, MeOH) indicated that the stereochemistry of 2 was the same as bionectin B [14]. Therefore, the structure of bionectin E was determined as shown.
Bionectin F (3) was obtained as a colorless oil and possessed a molecular formula of C50H96O8 as deduced from its HR-ESI-MS ([M+Na]+, m/z 847.7002) implying three degrees of unsaturation. The 1H and 13C NMR spectra exhibited three trisubstituted double bonds (δC 125.0 (d), 125.4 (d), 125.9 (d); 132.1 (s), 135.7 (s), 139.5 (s)) that attached to methylene (δH 5.36 (1H, t), 5.12 (1H, t), 5.08 (1H, t)), four allylic methyls (δH 1.66 (6H, s), 1.61 (3H, s), 1.59 (3H, s)), one allylic hydroxymethylene (δH 4.07 (2H, d), δC 59.4 (t)) [15]. For the structural characteristics of the polyprenols [15, 16], some oxygenated quaternary carbons, methylenes, and methyls signals were overlapped in 13C NMR spectra (Table 2 and Fig. S16). As the degrees unsaturation were all accounted for 3 double bonds, indicating the acyclic skeleton of 3. In 1H NMR spectrum, 88 proton signals were present (Table 2 and Fig. S15) and 8 protons overlapping with proton signals of CD3OD compare with its molecular formula, which is same with the numbers of oxygen atoms, requiring the existence of 8 OH groups in molecule. Moreover, the EI-MS fragments appeared as arithmetic sequence (Fig. S18), including I: 109, 177, 245, 313; II: 275, 343, 411, 479; III: 399, 467, 535; IV: 177, 263; V: 189, 275; VI: 245, 331; and VII: 313, 399. The differential between fragment groups I to III is 68 Da corresponding to a C5H8 and for groups IV–VII is 86 Da corresponding to a C5H10O, both of them are related to isoprene unit or its hydroxylate form. The above analysis showed that 3 possessed many characteristics of polyprenols and shared the same framework with polyprenols. Comparison with SCH 60061 [16], 3 shared same layout on the distribution of functional groups, including two double bonds contained isoprenes connected in head to tail fashion at the α-terminal and one residual double bond at the ω-terminal, but 3 contained 10 isoprene units that calculated from its molecular formula rather than 9 isoprene units in SCH 60061 [16]. This deduction was confirmed by the rational assignment of the detected EI-MS fragments (Fig. 4 and Fig. S18), which included two parts: one part included 135, 189, 275, 343, 411, and 479 peaks cleaving from the internal hydroxy isoprenes, another group contained 69, 109, 177, 263, 245, 331, 313, 399, 467, and 535 peaks cleaving from α-terminal E-isoprene [15]. The stereochemistry of the isoprene (CH2–C(CH3)=CH–CH2–) residues was determined to be all E-form by the 1H NMR chemical shifts of the allylic methyl signals (δH 1.66 (6H, s, H-40/41), 1.61 (3H, s, H-42)) [16, 17]. The configurations of other carbons cannot be determined by current data, so the structure of bionectin F (3) was determined as shown.
The known substances were identified as dioxopiperazine (4) [18], lasiodipline D (5) [19], glioclatine (6) [20], gliocladine B (7) [13], bionectin B (8) [14], 11,11′-dideoxyverticillin A (9) [18], verticillin A (10) [21], sch 52901 (11) [21], gliocladicillin A (12) [22], sch 52900 (13) [21], gliocladicillin C (14) [23], indole-3-acetic acid (15) [24], l-tryptophan (16) [25], cyclo(l-Pro- l-Leu) (17) [26], cerebroside C (18) [27], 2-furoic acid (19) [28], 2-hydroxy-5-hydroxymethyl furan (20) [29], 5-hydroxymethyl furoic acid (21) [28], and 5α,8α-epidioxyergosta-6,22-dien-3β-ol (22) [30] by comparison of their experimental and reported spectroscopic data.
The analogs of diketopiperazine and polyisoprenepolyol exhibited obviously antibacterial activity in previous investigation [14]. So the antibacterial activity of isolated compounds 1–3, 10, 11, and 14 were assayed against E. coli, Staphylococcus aureus, and Salmonella typhimurium ATCC 6539 with cefotaxime as a positive control. Compounds 1, 2, 10, 11, and 14 exhibited evident antibacterial activity (Table 3) and the MIC values were from 6.25–25 µg ml−1.
Materials and methods
General experimental procedures
The optical rotations were measured with a Jasco DIP-370 digital polarimeter. Ultraviolet (UV) spectra were recorded on a Shimadzu UV-2401PC spectrophotometer. The nuclear magnetic resonance (NMR) spectra were recorded on Avance III-600 spectrometers with tetramethylsilane (TMS) as an internal standard. The electrospray-ionization mass spectra (ESI-MS) and high-resolution electrospray-ionization mass spectra (HR-ESI-MS) were recorded on a Finnigan LCQ-Advantage mass spectrometer (Thermo, San Jose, USA) and a VG Auto-Spec-3000 mass spectrometer (VG, Manchester, UK), respectively. High resolution electron impact mass spectra (HR-EI-MS) were recorded on a Waters AutoSpec Premier P776 spectrometer electrospray ionization. Column chromatography was performed on silica gel 200-300 mesh, H, and GF254 (Qingdao Marine Chemical Inc., Qingdao, China) and Sephadex LH-20 (Amersham Pharmacia). Pre-coated silica gel GF254 plates (Qingdao Marine Chemical inc., Qingdao, China) were used for thin layer chromatography (TLC). Fractions were monitored by TLC and visualized by heating plates sprayed with 5% H2SO4 in EtOH.
Microbial strains, medium, and chemicals
The plant H. serrata was collected from Xichou County, Yunnan Province, People’s Republic of China, in July 2013. A voucher specimen (20130710ZPJ) was deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences. The isolation of fungus was done according to our previously reported protocol [8]. The isolated fungus strain Y1085 was identified by ITS sequencing, and the results (GenBank accession number MH429795.1) showed high homologies of 100% (in 512 bp) to Bionectria ochroleuca (GenBank accession number EU273558.1), which indicated that the strain Y1085 belongs to the genus Bionectria. The strain was deposited in State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, China.
Extraction and isolation of metabolites
The fermentation (10 l) of Bionectria sp. Y1085 was performed on petri dishes using solid yeast malt glucose (YMG) medium consisted of glucose 0.4%, yeast extract 0.4%, and malt extract 1% in distilled water, pH 7.5 at 26 °C for 14 days. The culture was then extracted exhaustively three times by EtOAc/MeOH/AcOH (80:15:5, v/v/v) to obtain the total crude extract (26.6 g) which was suspended in H2O and then extracted with EtOAc. The EtOAc extract (22.1 g) was applied on a column of silica gel CC eluted with a gradient of petroleum ether–EtOAc (25:1 to 3:2) firstly and then CHCl3–MeOH (100:3 to 0:1) to give six fractions (Fr.1 to Fr.6). Fr.2 (257.9 mg) was chromatographed over a silica gel column (200–300 mesh, 40 g) and eluted with CHCl3–MeOH with an increasing polarity that ranged from 200:1 to 100:2 to yield fractions Fr.2-1 to Fr.2-3. Fr.2-2 (67.8 mg) was purified by Sephadex LH-20 column of eluting with acetone to yield 22 (6.2 mg). Fr.2-3 (108.2 mg) was firstly loaded onto a column of Sephadex LH-20 as eluted with (CHCl3–MeOH, 1:1) to give fractions Fr.2-3-1 to Fr.2-3-6. Fr.2-3-4 and Fr.2-3-5 were subjected to semipreparative RP-C18 HPLC with a gradient elution of MeOH–H2O (15:85 to 100:0) and (10:90 to 100:0) for 30 min, respectively, to obtain 15 (1.4 mg) and 19 (8.3 mg). Fr.3 (4.9 g) was chromatographed over a silica gel column (200–300 mesh, 150 g) and eluted with CHCl3–MeOH (100:2 to 10:1) to give fractions Fr.3-1 to Fr.3-6. Fr.3-2 (3.8 g) was further chromatographed over a silica gel column (GF254, 60 g) and eluted with CHCl3–MeOH (100:0 to 100:8) to give fractions Fr.3-2-1 to Fr.3-2-7. Fr.3-2-2 (323.1 mg) was subjected to a silica gel GF254 column (30 g, CHCl3–acetone, 100:1) to obtain 11 (11.8 mg), 10 (18.0 mg) and 6 (2.9 mg). Fr.3-2-4 (2.3 g) was separated on a silica gel GF254 column (80 g, CHCl3–acetone, 100:1 to 100:8) to give fractions Fr.3-2-4-1 to Fr.3-2-4-12. Fr.3-2-4-3 (173.6 mg) and Fr.3-2-4-4 (643.6 mg) were subjected to a silica gel H and eluted with CHCl3–acetone (100:2 + 0.1% HCOOH), respectively, to obtain 14 (31.7 mg), 13 (59.2 mg), and 2 (2.4 mg). Fr.3-2-4-6 (9.6 mg) and Fr.3-2-4-8 (14.5 mg) were chromatographed over a silica gel GF254 and eluted with petroleum ether-acetone (7:3), respectively, to obtain compounds 7 (3.2 mg) and 9 (1.2 mg). Fr.3-2-4-10 (197.0 mg) was subjected to a silica gel GF254 column and eluted with CHCl3–acetone (7:3) to obtain compounds 12 (11.1 mg) and 17 (5.0 mg). Fr.3-2-5 (187.9 mg) was loaded onto a Sephadex LH-20 (CHCl3–MeOH, 1:1) and then further chromatographed over a silica gel GF254 (petroleum ether–acetone, 7:3) to obtain compound 8 (8.0 mg). Fr.3-3 (296.1 mg) was subjected on a silica gel GF254 eluting with CHCl3–acetone (9:1 to 7:3) to achieve compounds 4 (4.7 mg) and 5 (1.5 mg). Fr.3-6 (33.2 mg) was chromatographed over a silica gel GF254 and eluted with CHCl3–acetone (100:4 to 100:8 containing 0.1% HCOOH) to obtain compound 21 (5.4 mg). Fr.4 (1.6 g) was purified by Sephadex LH-20 (CHCl3–MeOH, 1:1) to give six fractions. Fr.4-3 (531.0 mg) was subjected to a silica gel GF254 and eluted with CHCl3–MeOH (25:1 to 7:3) to give seven fractions. Fr.4-3-2 (80.0 mg) was purified with Sephadex LH-20 (acetone) to obtain compound 1 (6.5 mg). Fr.4-3-7 (322.0 mg) was subjected to Sephadex LH-20 (MeOH) to obtain compound 20 (20.1 mg). Fr.4-4 (13.5 mg) was applied to a silica gel GF254 and eluted with CHCl3–MeOH (100:6 to 9:1) to obtain compound 16 (2.5 mg). Fr.5 (4.36 g) was purified with Sephadex LH-20 (CHCl3–MeOH, 1:1) to give fractions Fr.5-1 to Fr.5-5. Fr.5-1 (3.7 g) was subjected to a silica gel GF254 eluting with CHCl3–MeOH (25:1 to 3:2) to obtain compounds 3 (53.1 mg) and 18 (3.3 mg).
Bionectin D (1): Colorless amorphous powder; [α]\({\,\!}_{\mathrm{D}}^{17}\) + 117.0 (c 0.14, MeOH); UV (MeOH) λmax (log ε) 195 (4.59), 255 (4.57), 272 (3.81) nm; IR (KBr) cm−1: 3417 (OH and NH), 2923, 1671 (s, C=O); 1H NMR (CD3OD, 600 MHz) and 13C NMR (CD3OD, 150 MHz) data, see Table 1; positive ESI-MS m/z 356 [M+Na]+, 689 [2M+Na]+; HR-EI-MS m/z 333.1148 [M]+ (calcd for C16H19N3O3S, 333.1147).
Bionectin E (2): White powder; [α]\({\,\!}_{\mathrm{D}}^{24}\) + 417.4 (c 0.14, MeOH); UV (MeOH) λmax (log ε) 203 (4.59), 218 (4.57), 281 (3.81) nm; IR (KBr) cm−1: 3414 (OH and NH), 2920, 1673 (s, C=O); 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data, see Table 1; positive ESI-MS m/z 479 [M+H]+, 501 [M+Na]+, 979 [2M+Na]+; HR-ESI-MS m/z 501.1024 [M+Na]+ (calcd for C24H22N4O3S2Na, 501.1026).
Bionectin F (3): Colorless oil; [α]\({\,\!}_{\mathrm{D}}^{17}\) − 11.9 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 195 (2.82) nm; IR (KBr) cm−1: 3388 (OH), 2911, 1463, 1375, 1177, 936; 1H NMR (CD3OD, 600 MHz) and 13C NMR (CD3OD, 150 MHz) data, see Table 2; positive ESI-MS m/z 825 [M+H]+, 847 [M+Na]+; HR-ESI-MS m/z 847.7002 [M+Na]+ (calcd for C50H96O8Na, 847.7003); EI-MS m/z 716 (2), 698 (4), 680 (4), 535 (4), 479 (5), 467 (6), 411 (8), 399 (6), 343 (9), 331 (7), 313 (4), 275 (9), 263 (4), 245 (7), 189 (20), 177 (22), 135 (36), 109 (54), 69 (85).
Assay anti-bacterial activity
The MIC values of isolated compounds 1–3, 10, 11, and 14 against E. coli, Staphylococcus aureus, and Salmonella typhimurium ATCC 6539 were determined by standard microdilution plate method, as described previously [9]. Briefly, bacteria were firstly incubated in nutrient agar at 25 °C for 18 h, and then they were introduced into sterile 0.9% saline solution with a cell suspension of 106 colony-forming units/ml (CFU/ml) for assay. The tested compound solution (100 μl) with series of concentrations were added, respectively, into 96-well plates containing bacteria suspension (100 μl) to make sure the final concentration of sample ranging from 0.78–100 μg ml−1, and then the plates were incubated at 37 °C for 24 h. After that, the wells were examined for growth of bacteria by measuring optical density (OD) values. All of the experiments were performed in triplicate.
References
Saleem M, Ali MS, Hussain S, Jabbar A, Ashraf M, Lee YS. Marine natural products of fungal origin. Nat Prod Rep. 2007;24:1142–52.
Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J Nat Prod. 2004;67:257–68.
Stringlis IA, Zhang H, Pieterse CMJ, Bolton MD, de Jonge R. Microbial small molecules–weapons of plant subversion. Nat Prod Rep. 2018;35:410–33.
Zhang HW, Song YC, Tan RX. Biology and chemistry of endophytes. Nat Prod Rep. 2006;23:753–71.
Kharwar RN, Mishra A, Gond SK, Stierle A, Stierle D. Anticancer compounds derived from fungal endophytes: their importance and future challenges. Nat Prod Rep. 2011;28:1208–28.
Kaul S, Gupta S, Ahmed M, Dhar MK. Endophytic fungi from medicinal plants: a treasure hunt for bioactive metabolites. Phytochem Rev. 2012;11:487–505.
Ferreira A, Rodrigues M, Fortuna A, Falcao A, Alves G. Huperzine A from Huperzia serrata: a review of its sources, chemistry, pharmacology and toxicology. Phytochem Rev 2016;15:51–85.
Chen YM, Yang YH, Li XN, Zou C, Zhao PJ. Diterpenoids from the endophytic fungus Botryosphaeria sp. P483 of the chinese herbal medicine Huperzia serrata. Molecules. 2015;20:16924–32.
Yu FX, Chen Y, Yang YH, Zhao PJ. Four new dimeric spiro-azaplilone derivatives cochliodones E-H from the entophytic fungus Chaetomium sp. M336. Phytochem Lett. 2016;16:263–7.
Zheng CJ, Kim YH, Kim WG. Glioperazine B, as a new antimicrobial agent against Staphylococcus aureus, and glioperazine C: two new dioxopiperazines from Bionectra byssicola. Biosci Biotechnol Biochem. 2007;71:1979–83.
Usami Y, Yamaguchi J, Numata A, Gliocladins A–C. and glioperazine: cytotoxic dioxo– or trioxopiperazine metabolites from a Gliocladium sp. separated from a sea hare. Heterocycles. 2004;63:1123–9.
Zhao WY, et al. Three new dioxopiperazine metabolites from a marine-derived fungus Aspergillus fumigatus Fres. Nat Prod Res. 2010;24:953–7.
Dong JY, He HP, Shen YM, Zhang KQ. Nematicidal epipolysulfanyldioxopiperazines from Gliocladium roseum. J Nat Prod. 2005;68:1510–3.
Zheng CJ, Kim CJ, Bae KS, Kim YH, Kim WG, Bionectins A-C. epidithiodioxopiperazines with anti-MRSA activity, from Bionectra byssicola F120. J Nat Prod. 2006;69:1816–19.
Nishida H, Huang XH, Tomoda H, Omura S. Glisoprenins, new inhibitors of acyl-CoA: cholesterol acyltransferase produced by Gliocladium sp. FO-1513. II. Structure elucidation of glisoprenins A and B. J Antibiot. 1992;45:1669–76.
Hegde VR, et al. Neurokinin receptor inhibitors: fermentation, isolation, physico-chemical properties, structure and biological activity. J Antibiot. 1997;50:983–91.
Chu CY, Vukov R. Determination of the structure of butyl rubber by NMR spectroscopy. Macromolecules. 1985;18:1423–30.
Son BW, Jensen PR, Kauffman CA, Fenical W. New cytotoxic epidithiodioxopiperazines related to verticillin A from a marine isolate of the fungus Penicillium. Nat Prod Lett. 1999;13:213–22.
Wei W, et al. An antibacterial metabolite from Lasiodiplodia pseudotheobromae F2. Phytochemistry. 2014;100:103–9.
Dong JY, Zhou W, Li L, Li GH, Liu YJ, Zhang KQ. A new epidithiodioxopiperazine metabolite isolated from Gliocladium roseum YMF1.00133. Chin Chem Lett. 2006;17:922–4.
Chu M, et al. Inhibition of c-fos proto-oncogene induction by Sch 52900 and Sch 52901, novel diketopiperazine produced by Gliocladium sp. J Antibiot. 1995;48:1440–45.
Chen Y, Guo H, Du Z, Liu XZ, Che Y, Ye X. Ecology-based screen identifies new metabolites from a Cordyceps-colonizing fungus as cancer cell proliferation inhibitors and apoptosis inducers. Cell Prolif. 2009;42:838–47.
Che YS, Liu XZ, Ye X, Guo HJ, Chen YL, Liu SC. Method for preparing gliocladicillin C and application thereof. CN101805699, no. 2009-10077302-101805699. 2009;February:17.
Evidente A, Iacobellis NS, Sisto A. Isolation of indole-3-acetic acid methyl ester, a metabolite of indole-3-acetic acid from Pseudomonas amygdali. Experientia. 1993;49:182–3.
Yan XJ, Suzuki M, Ohnishi-Kameyama M, Sada Y, Nakanishi T, Nagata T. Extraction and identification of antioxidants in the roots of Yacon (Smallanthus sonchifolius). J Agric Food Chem. 1999;47:4711–3.
Zhao PJ, Wang HX, Li GH, Li HD, Liu J, Shen YM. Secondary metabolites from endophytic Streptomyces sp. Lz531. Chem Biodivers. 2007;4:899–904.
Koga J, et al. Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J Biol Chem. 1998;273:31985–91.
Yang T, Wang CH, Liu HJ, Chou GX, Cheng XM, Wang ZT. A new antioxidant compound from Capparis spinose. Pharm Biol. 2010;48:589–94.
Saleem R, Ahmad M, Naz A, Siddiqui H, Ahmad SI, Faizi S. Hypertensive and toxicological study of citric acid and other constituents from Tagetes patula roots. Arch Pharm Res. 2004;27:1037–42.
Yue JM, Chen SN, Lin ZW, Sun HD. Sterols from the fungus Lactarium volemus. Phytochemistry. 2001;56:801–6.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (31760018, 31560016), the Applied Basic Research Foundation of Yunnan Province (2018FA006 and 2017FA016), and National Key R&D Program of China (2017YFD0201104).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, YH., Yang, DS., Li, GH. et al. Antibacterial diketopiperazines from an endophytic fungus Bionectria sp. Y1085. J Antibiot 72, 752–758 (2019). https://doi.org/10.1038/s41429-019-0209-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-019-0209-5
This article is cited by
-
Antitumor and antibacterial activity of metabolites of endophytic Colletotrichum siamense isolated from coffee (Coffea arabica L. cv IAPAR-59)
Brazilian Journal of Microbiology (2023)