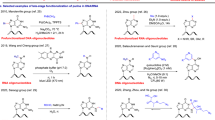
Abstract
Xenonucleic acids (XNAs), which are composed of artificial scaffolds and natural nucleobases, have unique hybridization properties that depend on the scaffold structure. Here, we functionalized the acyclic XNA serinol nucleic acid (SNA) with nonnatural nucleobases. A linear SNA probe functionalized with 5-perylenylethynyl uracil residues showed weak greenish-yellow excimer emission in the absence of target RNA and bright cyan-green monomer emission in the presence of target RNA. Probe hybridization was rapid and enabled the quantitative measurement of RNA with discrimination of single-base mismatches. We also designed a photoresponsive SNA with two 8-pyrenylvinyl adenine (PVA) residues. Irradiation with blue (455 nm) light caused [2 + 2] photocycloaddition between intrastrand PVAs, resulting in the dissociation of the SNA/RNA duplex, whereas irradiation with ultraviolet (340 nm) light induced cycloreversion of the PVA photodimer and SNA/RNA duplex reformation. Using a combination of 8-naphthylvinyl adenine (NVA) and PVA and irradiation with 465 nm, 405 nm, 340 nm, and 300 nm light, orthogonal control of the formation of SNA(NVA-NVA)/RNA and SNA(PVA-NVA)/RNA duplexes was demonstrated. Thus, nucleobase modifications further expand the utility of acyclic XNA in bionanotechnology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Beaucage SL, Iyer RP. Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron. 1992;48:2223–311.
Leumann CJ. DNA analogues: from supramolecular principles to biological properties. Bioorg Med Chem. 2002;10:841–54.
Zhang S, Switzer C, Chaput JC. The resurgence of acyclic nucleic acids. Chem Biodivers. 2010;7:245–58.
Pinheiro VB, Holliger P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr Opin Chem Biol. 2012;16:245–52.
Krishnamurthy R. On the emergence of RNA. Isr J Chem. 2015;55:837–50.
Chaput JC, Herdewijn P. What Is XNA? Angew Chem Int Ed. 2019;58:11570–2.
Murayama K, Asanuma H. Design and hybridization properties of acyclic xeno nucleic acid oligomers. ChemBioChem. 2021;22:2507–15.
Asanuma H, Kamiya Y, Kashida H, Murayama K. Xeno nucleic acids (XNAs) having non-ribose scaffolds with unique supramolecular properties. Chem Commun. 2022;28:3993.
Hövelmann F, Seitz O. DNA stains as surrogate nucleobases in fluorogenic hybridization probes. Acc Chem Res. 2016;49:714–23.
Samanta D, Ebrahimi SB, Mirkin CA. Nucleic-acid structures as intracellular probes for live cells. Adv Mater. 2020;32:1901743.
Vilaivan T. Fluorogenic PNA probes. Beilstein J Org Chem. 2018;14:253–81.
Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–8.
Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–64.
Le BT, Chen S, Abramov M, Herdewijn P, Veedu RN. Evaluation of anhydrohexitol nucleic acid, cyclohexenyl nucleic acid and D-altritol nucleic acid-modified 2′-O-methyl RNA mixmer antisense oligonucleotides for exon skipping in vitro. Chem Commun. 2016;52:13467–70.
McClorey G, Moulton HM, Iversen PL, Fletcher S, Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–81.
Laursen MB, Pakula MM, Gao S, Fluiter K, Mook OR, Baas F, et al. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol Biosyst. 2010;6:862.
Takahashi M, Nagai C, Hatakeyama H, Minakawa N, Harashima H, Matsuda A. Intracellular stability of 2′-OMe-4′-thioribonucleoside modified siRNA leads to long-term RNAi effect. Nucleic Acids Res. 2012;40:5787–93.
Schlegel MK, Foster DJ, Kel’in AV, Zlatev I, Bisbe A, Jayaraman M, et al. Chirality dependent potency enhancement and structural impact of glycol nucleic acid modification on siRNA. J Am Chem Soc. 2017;139:8537–46.
Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol. 2017;35:238–48.
Yu H, Zhang S, Chaput JC. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem. 2012;4:183–7.
Taylor AI, Pinheiro VB, Smola MJ, Morgunov AS, Peak-Chew S, Cozens C, et al. Catalysts from synthetic genetic polymers. Nature. 2015;518:427–30.
Eremeeva E, Fikatas A, Margamuljana L, Abramov M, Schols D, Groaz E, et al. Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acids Res. 2019;47:4927–39.
Hoshino H, Kasahara Y, Kuwahara M, Obika S. DNA polymerase variants with high processivity and accuracy for encoding and decoding locked nucleic acid sequences. J Am Chem Soc. 2020;142:21530–7.
Asanuma H, Toda T, Murayama K, Liang X, Kashida H. Unexpectedly stable artificial duplex from flexible acyclic threoninol. J Am Chem Soc. 2010;132:14702–3.
Murayama K, Kashida H, Asanuma H. Acyclic L-threoninol nucleic acid (L-aTNA) with suitable structural rigidity cross-pairs with DNA and RNA. Chem Commun. 2015;51:6500–3.
Murayama K, Kashida H, Asanuma H. Methyl group configuration on acyclic threoninol nucleic acids (aTNAs) impacts supramolecular properties. Org Biomol Chem. 2022;20:4115–22.
Kashida H, Murayama K, Toda T, Asanuma H. Control of the chirality and helicity of oligomers of serinol nucleic acid (SNA) by sequence design. Angew Chem Int Ed. 2011;50:1285–8.
Murayama K, Kamiya Y, Kashida H, Asanuma H. Ultrasensitive molecular beacon designed with totally serinol nucleic acid (SNA) for monitoring mRNA in cells. ChemBioChem. 2015;16:1298–301.
Le BT, Murayama K, Shabanpoor F, Asanuma H, Veedu RN. Antisense oligonucleotide modified with serinol nucleic acid (SNA) induces exon skipping in mdx myotubes. RSC Adv. 2017;7:34049–52.
Asanuma H, Murayama K, Kamiya Y, Kashida H. The DNA duplex as an aqueous one-dimensional soft crystal scaffold for photochemistry. Bull Chem Soc Jpn. 2018;91:1739–48.
Asanuma H, Murayama K, Kamiya Y, Kashida H. Design of photofunctional oligonucleotides by copolymerization of natural nucleobases with base surrogates prepared from acyclic scaffolds. Polym J. 2017;49:279–89.
Abdelhady AM, Onizuka K, Ishida K, Yajima S, Mano E, Nagatsugi F. Rapid alkene–alkene photo-cross-linking on the base-flipping-out field in duplex DNA. J Org Chem. 2022;87:2267–76.
Morihiro K, Kodama T, Waki R, Obika S. Light-triggered strand exchange reaction using the change in the hydrogen bonding pattern of a nucleobase analogue. Chem Sci. 2014;5:744–50.
Barrois S, Wagenknecht HA. Diarylethene-modified nucleotides for switching optical properties in DNA. Beilstein J Org Chem. 2012;8:905–14.
Cahová H, Jäschke A. Nucleoside-based diarylethene photoswitches and their facile incorporation into photoswitchable DNA. Angew Chem Int Ed. 2013;52:3186–90.
Ogasawara S, Maeda M. Straightforward and reversible photoregulation of hybridization by using a photochromic nucleoside. Angew Chem Int Ed. 2008;47:8839–42.
Wada T, Minamimoto N, Inaki Y, Inoue Y. Peptide Ribonucleic Acids (PRNA). 2. A Novel Strategy for Active Control of DNA Recognition through Borate Ester Formation. J Am Chem Soc. 2000;122:6900–10.
Ohkubo A, Kasuya R, Miyata K, Tsunoda H, Seio K, Sekine M. New thermolytic carbamoyl groups for the protection of nucleobases. Org Biomol Chem. 2009;7:687.
Scharf P, Müller J. Nucleic acids with metal-mediated base pairs and their applications. Chempluschem. 2013;78:20–34.
Liu Q, Deiters A. Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Acc Chem Res. 2014;47:45–55.
Dietzsch J, Bialas D, Bandorf J, Würthner F, Höbartner C. Tuning exciton coupling of merocyanine nucleoside dimers by RNA, DNA and GNA double helix conformations. Angew Chem Int Ed. 2022;61:e202116783.
Seo YJ, Rhee H, Joo T, Kim BH. Self-duplex formation of an A Py-substituted oligodeoxyadenylate and its unique fluorescence. J Am Chem Soc. 2007;129:5244–7.
Okamoto A, Kanatani K, Saito I. Pyrene-labeled base-discriminating fluorescent DNA probes for homogeneous SNP typing. J Am Chem Soc. 2004;126:4820–7.
Skorobogatyi MV, Malakhov AD, Pchelintseva AA, Turban AA, Bondarev SL, Korshun VA. Fluorescent 5-Alkynyl-2′-Deoxyuridines: High Emission Efficiency of a Conjugated Perylene Nucleoside in a DNA Duplex. ChemBioChem. 2006;7:810–6.
Börjesson K, Preus S, El-Sagheer AH, Brown T, Albinsson B, Wilhelmsson LM. Nucleic Acid Base Analog FRET-Pair Facilitating Detailed Structural Measurements in Nucleic Acid Containing Systems. J Am Chem Soc. 2009;131:4288–93.
Han JH, Yamamoto S, Park S, Sugiyama H. Development of a Vivid FRET System Based on a Highly Emissive dG-dC Analogue Pair. Chem A Eur J. 2017;23:7607–13.
Okamura H, Trinh GH, Dong Z, Masaki Y, Seio K, Nagatsugi F. Selective and stable base pairing by alkynylated nucleosides featuring a spatially-separated recognition interface. Nucleic Acids Res. 2022;50:3042–55.
Morihiro K, Moriyama Y, Nemoto Y, Osumi H, Okamoto A. Anti–syn Unnatural Base Pair Enables Alphabet-Expanded DNA Self-Assembly. J Am Chem Soc. 2021;143:14207–17.
Takezawa Y, Suzuki A, Nakaya M, Nishiyama K, Shionoya M. Metal-Dependent DNA Base Pairing of 5-Carboxyuracil with Itself and All Four Canonical Nucleobases. J Am Chem Soc. 2020;142:21640–4.
Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 2009;37:e14–e14.
Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE. PCR with an Expanded Genetic Alphabet. J Am Chem Soc. 2009;131:14620–1.
Kaul C, Müller M, Wagner M, Schneider S, Carell T. Reversible bond formation enables the replication and amplification of a crosslinking salen complex as an orthogonal base pair. Nat Chem. 2011;3:794–800.
St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–75.
Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci. 2003;100:13308–13.
Asanuma H, Akahane M, Kondo N, Osawa T, Kato T, Kashida H. Quencher-free linear probe with multiple fluorophores on an acyclic scaffold. Chem Sci. 2012;3:3165.
Murayama K, Asanuma H. A Quencher‐Free Linear Probe from Serinol Nucleic Acid with a Fluorescent Uracil Analogue. ChemBioChem. 2020;21:120–8.
Brieke C, Rohrbach F, Gottschalk A, Mayer G, Heckel A. Light‐Controlled Tools. Angew Chem Int Ed. 2012;51:8446–76.
Lubbe AS, Szymanski W, Feringa BL. Recent developments in reversible photoregulation of oligonucleotide structure and function. Chem Soc Rev. 2017;46:1052–79.
Yoshimura Y, Fujimoto K. Ultrafast Reversible Photo-Cross-Linking Reaction: Toward in Situ DNA Manipulation. Org Lett. 2008;10:3227–30.
Asanuma H, Liang X, Nishioka H, Matsunaga D, Liu M, Komiyama M. Synthesis of azobenzene-tethered DNA for reversible photo-regulation of DNA functions: hybridization and transcription. Nat Protoc. 2007;2:203–12.
Goldau T, Murayama K, Brieke C, Steinwand S, Mondal P, Biswas M, et al. Reversible Photoswitching of RNA Hybridization at Room Temperature with an Azobenzene C -Nucleoside. Chem - A Eur J. 2015;21:2845–54.
Škugor M, Valero J, Murayama K, Centola M, Asanuma H, Famulok M. Orthogonally Photocontrolled Non‐Autonomous DNA Walker. Angew Chem Int Ed. 2019;58:6948–51.
Kamiya Y, Asanuma H. Light-Driven DNA Nanomachine with a Photoresponsive Molecular Engine. Acc Chem Res. 2014;47:1663–72.
Wenge U, Wengel J, Wagenknecht HA. Photoinduced Reductive Electron Transfer in LNA:DNA Hybrids: A Compromise between Conformation and Base Stacking. Angew Chem Int Ed. 2012;51:10026–9.
Zhang RS, McCullum EO, Chaput JC. Synthesis of Two Mirror Image 4-Helix Junctions Derived from Glycerol Nucleic Acid. J Am Chem Soc. 2008;130:5846–7.
Hsieh WC, Martinez GR, Wang A, Wu SF, Chamdia R, Ly DH. Stereochemical conversion of nucleic acid circuits via strand displacement. Commun Chem. 2018;1:89.
Kim KT, Angerani S, Winssinger N. A minimal hybridization chain reaction (HCR) system using peptide nucleic acids. Chem Sci. 2021;12:8218–23.
Murayama K, Yamano Y, Asanuma H. 8-Pyrenylvinyl Adenine Controls Reversible Duplex Formation between Serinol Nucleic Acid and RNA by [2 + 2] Photocycloaddition. J Am Chem Soc. 2019;141:9485–9.
Kovalenko NP, Abdukadyrov AT, Gerko VI, Alfimov MV. Luminescent and photochemical behaviour of diarylethylenes with 3-pyrenyl substituents. J Photochem. 1980;12:59–65.
Rodrigues-Correia A, Weyel XMM, Heckel A. Four Levels of Wavelength-Selective Uncaging for Oligonucleotides. Org Lett. 2013;15:5500–3.
Fujimoto K, Sasago S, Mihara J, Nakamura S. DNA Photo-cross-linking Using Pyranocarbazole and Visible Light. Org Lett. 2018;20:2802–5.
Nishioka H, Liang X, Kato T, Asanuma H. A Photon-Fueled DNA Nanodevice that Contains Two Different Photoswitches. Angew Chem Int Ed. 2012;51:1165–8.
Haydell MW, Centola M, Adam V, Valero J, Famulok M. Temporal and Reversible Control of a DNAzyme by Orthogonal Photoswitching. J Am Chem Soc. 2018;140:16868–72.
Yamano Y, Murayama K, Asanuma H. Dual Crosslinking Photo‐Switches for Orthogonal Photo‐Control of Hybridization Between Serinol Nucleic Acid and RNA. Chem – A Eur J. 2021;27:4599–604.
Murata S, Toyota T, Nomura SM, Nakakuki T, Kuzuya A. Molecular Cybernetics: Challenges toward Cellular Chemical Artificial Intelligence. Adv Funct Mater. 2022;32:2201866.
Kamiya Y, Donoshita Y, Kamimoto H, Murayama K, Ariyoshi J, Asanuma H. Introduction of 2,6-Diaminopurines into Serinol Nucleic Acid Improves Anti-miRNA Performance. ChemBioChem. 2017;18:1917–22.
Sato F, Kamiya Y, Asanuma H. Syntheses of Base-Labile Pseudo-Complementary SNA and L-aTNA Phosphoramidite Monomers. J Org Chem. 2023. https://doi.org/10.1021/acs.joc.2c01911.
Kashida H, Hattori Y, Tazoe K, Inoue T, Nishikawa K, Ishii K, et al. Bifacial Nucleobases for Hexaplex Formation in Aqueous Solution. J Am Chem Soc. 2018;140:8456–62.
Chen Y, Nagao R, Murayama K, Asanuma H. Orthogonal Amplification Circuits Composed of Acyclic Nucleic Acids Enable RNA Detection. J Am Chem Soc. 2022;144:5887–92.
Acknowledgements
This work was supported by a Grant-in-Aid for Transformative Research Areas “Molecular Cybernetics” JP20H05970 (KM), 20H05968 (KM), JSPS KAKENHI grants JP20K15399 (KM), JP22J01195 (YY), JP22K14792 (YY), and JP21H05025 (HA). AMED under Grant Number 22am0401007 (HA) is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Murayama, K., Yamano, Y. & Asanuma, H. Functionalization of acyclic xenonucleic acid with modified nucleobases. Polym J 55, 743–752 (2023). https://doi.org/10.1038/s41428-023-00776-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41428-023-00776-7