Abstract
Amid the COVID-19 pandemic, cancer continues to be the most devastating disease worldwide. Liquid biopsy of circulating tumor cells (CTCs) has recently become a painless and noninvasive tool for obtaining carcinoma cell samples for molecular profiling. Here, we report efficient detection and collection of cancer cells in blood samples by combining stem cell antigen (CD44)-specific immunosilica particles and immunofluorescent quantum dots with spectrally and temporally resolved single-photon counting. We accurately detect 1–10 cells among 100 cancer cells of the breast, lungs, or cervix in 1 mL blood samples. In addition, the bright and narrowband emission of CdSe/ZnS quantum dots enables temporally and spectrally resolved photon counting for multiplexed cancer cell detection. The cancer cell-specific and large immunosilica particles helped us collect the specific cells. We validate the detection efficiency and multimodality of this strategy by time-stamped and energy-dispersed single-photon counting of orange- and red-emitting quantum dots and green-fluorescing nuclei stained with Syto-13/25 dye. Thus, the present work highlights the prospects of multimodal CTC detection for noninvasive cancer screening and postsurgical or therapeutic follow-up.
Similar content being viewed by others
Introduction
COVID-19 and cancer are the major life-threatening challenges facing humanity today. Although cooperative research at the molecular biology, immunology, and genetics interface has controlled the COVID-19 pandemic1,2,3, cancer continues to be the most devastating disease worldwide, underscoring the significance of advanced cancer screening and therapeutic modalities4,5,6,7. Liquid biopsy of metastatic cells or circulating tumor cells (CTCs) have shown their excellence in the isolation, early stage detection, and genomic identification of various carcinomas4,8,9,10,11,12. CTCs invade the circulatory or lymphatic systems after shedding from the parent tumor, eventually causing metastasis13,14,15. Hence, CTCs are potential targets for cancer management by providing crucial therapeutic information to ascertain cancer recurrence and enhance chemotherapeutic outcomes16,17,18,19. Nevertheless, the extreme rarity of CTCs calls for accurate isolation and enumeration strategies20,21.
Fluorescence is a promising tool for CTC detection22,23, for which the collection of cells from a blood sample is essential. Overexpressed epithelial antigens on CTCs are detectable targets. Antigen-specific antibodies (abs) conjugated to nanomaterials such as graphene oxide24, silicone nanopillars25, gold nanoparticles26, magnetosomes27, Strep tags28, silica particles29,30,31, or quantum dots (QDs)32,33,34,35 are promising tags for CTC detection. These abs include anti-EpCAM ab10,20, anti-CD44 ab12, anti-EGFR ab36, anti-HER2 ab37, and anti-Vimentin ab38. However, the epithelial-to-mesenchymal transition (EMT) in CTCs downregulates the epithelial phenotypes such as EpCAM, making accurate CTC detection challenging39,40. Recent studies have shown the association of stem cell antigens and the CD44 expressing phenotype with many metastatic tumors and CTCs41,42. Furthermore, most CTC detection procedures targeting anticancer antigens on epithelial cells show single modality enrichment, limiting the CTC detection accuracy of samples obtained by liquid biopsy.
We collected cancer cells (MCF7 and HeLa cells) from blood samples using immunosilica microparticles targeting the mesenchymal antigen CD44. The large silica particle size helped us separate cancer cells without the use of a magnetic or centrifugal force. Additionally, the large particle size prevents endocytosis commonly observed for silica or magnetic nanoparticles. We accomplished error-free characterization of collected CD44-specific cancer cells by performing triple fluorescence staining, multicolor imaging, and deconvoluting the fluorescence spectral and decay profile data. Triple staining of MCF7 and HeLa cells was performed using Syto-13/25 nucleus-staining dye, a CdSe/ZnS quantum dot (QD585)-conjugated anti-EpCAM ab, and a QD655-conjugated anti-CD44 ab. Similarly, we isolated and characterized H1650 and HeLa cells in blood samples using anti-EGFR-ab-conjugated silica particles.
Experimental section
Materials and reagents
Mesoporous silica particles (FUJIFILM Wako), streptavidin (FUJIFILM Wako), phosphate-buffered saline (PBS, FUJIFILM Wako), (3-aminopropyl)triethoxysilane (APTES, TCI, Japan), (D-/L-) biotin N-hydroxysuccinimide ester (biotin-NHS, Sigma, Japan), anti-EGFR ab (Sigma, Japan), biotin-anti-EpCAM ab (BioLegend, USA), anti-CD44 ab (Sony, Japan), Alexa Fluor®488 anti-CD44 ab (Sony, Japan), streptavidin-QD585/655-conjugate (Thermo Fisher Scientific, USA), fetal bovine serum (FBS, Thermo Fisher Scientific, USA), penicillin/streptomycin (P/S, Thermo Fisher Scientific, USA), trypsin (Thermo Fisher Scientific, USA), and Syto-13/25 (Thermo Fisher Scientific, USA) were used as obtained.
Preparation of ab-functionalized silica microparticles
Mesoporous silica microparticles (2.0 mg; 75 ~ 150 µm size) were silanized by adding 25 µL of APTES solution (1 wt% APTES, 80 wt% acetone, and 19 wt% water). SEM images showing the surface morphologies of the silica particles are presented in Fig. S1. Silanization was carried out in a microtube in a shaker for 30 min (25 °C). The settled particles were thoroughly rinsed with acetone and PBS and stabilized in PBS. Biotin-NHS (10 eq.) dissolved in PBS was then added to the silanized microparticles, allowed to react at RT for 30 min by gently mixing, and washed with PBS and water. Next, a streptavidin solution (12.5 eq.) in PBS was added to the biotinylated microparticles and reacted at RT for 30 min, similarly to the previous step, followed by washing with PBS. A separate solution of biotinylated anti-CD44 ab (30 eq.) was prepared in PBS. It was added to the streptavidin-functionalized silica microparticles, reacted at 25 °C (30 min), and continuously mixed by up-down flipping. The microparticle-ab conjugate sample was rinsed with PBS, stabilized in PBS, and stored at 4 °C. Additionally, a monoclonal anti-EGFR-ab-silica conjugate was prepared as explained above.
Cell culture, labeling, and detection
MCF7 and HeLa cells were cultured in DMEM with 10% FBS and 1% P/S. NCI-H1650 cells and human T cells were cultured in RPMI 1640 medium with 100 U/mL P/S, 250 ng/mL fungizone, and 10% FBS and collected by centrifugation.
MCF7 or HeLa cells (~70% confluence) were incubated with QD655-anti-EpCAM-ab and QD585-anti-CD44-ab conjugates (6 nM, 37 °C, 30 min). The cells were washed with PBS and incubated with Syto-13 nucleus-staining dye (250 nM) for 15 min. The antigen expression level was determined from fluorescence photocount images in a 200 ns window with 500 to 800 nm spectral resolution. An optical parametric amplifier (OPA) was pumped by a regenerative amplifier (RegA) to generate 405 nm (200 kHz, 150 fs) output pulses as the excitation light source. An oscillator (76 MHz) seeded the RegA. Single-photon maps were generated using a polychromator-streak camera assembly. The fluorescence signals (>500 nm) were identified using a filter, and the spectrally and temporally resolved photons from the polychromator were counted with the streak camera. PL lifetime values were calculated by fitting PL decay values to the third-exponential equation. Fluorescence images were taken with an Olympus microscope (IX70) through a 10X or 40X objective lens and a 500 nm longpass filter. The labeled cells were excited with a 465 nm (200 mW) continuous wave (cw) laser (ACAN A12), and fluorescence images were obtained using a digital camera (OLYMPUS, Model No. XZ-2).
Blood sample collection and PBMC isolation
We collected and used PBMCs following the procedure approved by the Ethics Committee (No. 2020–7) of the Graduate School of Dental Medicine, Hokkaido University. PBMCs from a healthy adult donor were collected and isolated by centrifugation with Histopaque-1077 (Merck, USA) according to the manufacturer’s protocol. Briefly, 3 mL of whole blood was layered on top of 3 mL of Histopaque-1077. After centrifugation, the plasma layer was discarded, and the layer containing mononuclear cells was collected. The cells were washed with the cell culture medium and resuspended in the cell culture medium. This step was repeated twice, and finally, the cells were mixed well by gentle pipetting.
Capture and identification of CTCs in a model blood sample
MCF7 or HeLa cells were spiked into a solution of PBMCs in a buffer resembling the clinical samples. The cancer cell concentrations were set to 104, 103, or 102 cells/mL, and the PBMC number was maintained at a105 cells in all cases. We applied two strategies for cell labeling. First, we obtained unlabeled cancer cells with anti-EpCAM-ab-conjugated silica particles and then stained the cells with Syto-13/25 and QD585/655 ab conjugates. However, we found intense background fluorescence due to the nonspecific binding of the QDs to the silica (Fig. S2), which prevented us from efficiently detecting the cancer cells attached to the particles. Therefore, to eliminate background noise, we followed a pre-labeling procedure. In this procedure, cells in a blood sample were labeled with Syto-13 nucleus-staining dye (250 nM), QD655-anti-EpCAM ab (6 nM), and QD585-anti-CD44-ab conjugates (6 nM) by coincubation (25 °C, 30 min). The cells were collected, washed with PBS, and resuspended in PBS. The prelabeled cells were separately added to modified anti-CD44-ab-conjugated silica microparticles (1000 particles/mL). The cells were collected on silica particles by continuously flipping the silica-cell suspension up and down for 30 min, and then, the silica particles settled under gravity. The settled particles were gently washed with PBS to minimize the detachment of the cells. The particles were dispersed on a glass slide, mounted on the microscope, and excited with a 405 nm fs laser. Temporally and spectrally resolved photocount maps were obtained using a polychromator-Streak camera assembly. Additionally, a color CCD camera was used to obtain multicolor images of the cells attached to the immunosilica particles.
To generalize the cell collection method, we repeated the aforementioned experiments using silica microparticles functionalized with an anti-EGFR ab to identify EGFR-overexpressing carcinoma cells such as H1650 or HeLa and used T cells as modal blood cells. In this experiment, fluorescence staining of HeLa cells was carried out using a QD655-anti-EpCAM-ab conjugate (6 nM), AlexaFluor488 anti-CD44-ab conjugate (6 nM), and Syto-25 (250 nM), whereas H1650 and T cells were stained only with Syto-25.
Results and discussion
Determination of CD44 and EpCAM antigen levels in MCF7 cells
We examined the labeling efficiency of MCF7 cells, the most common breast cancer cells, with anti-CD44 ab and anti-EpCAM ab43. Briefly, cultured MCF7 cells were labeled by incubation with Syto-13 nucleus-staining dye, QD585-anti-CD44 ab, and QD655-anti-EpCAM ab. The details of the cell culture condition, ab conjugate preparation, and cell labeling process are presented in the experimental section. Figure 1A shows a fluorescence image of the immunostained cells excited at 465 nm and measured with a 480-nm longpass filter. Here, the number of CD44 and EpCAM antigens expressed on the cell surface was determined by time-resolved fluorescence photon counting. The photons emitted from the triply stained cells were identified using a spectrometer-streak camera system.
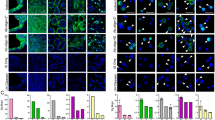
A Fluorescence image of the cells showing the emission from Syto-13 (green), QD585-anti-CD44 ab (orange), and QD655-anti-EpCAM ab (red). B A single-photon histogram of the cells. The lower part of the histogram in B shows the fluorescence spectra (approximately 515, 585, and 655 nm) deconvoluted from the histogram data. The broken lines represent the Lorentzian fits based on the three fluorescence intensity maxima. C The fluorescence decay curves deconvoluted from the data in the histogram in B.
We characterized MCF7 cells using a three-color fluorescence image and a single-photon histogram (Fig. 1B). The histogram was further resolved by deconvoluting the energy-dispersed fluorescence photons (Fig. 1B) and generating three decay curves (Fig. 1C) with three distinct fluorescence lifetime values (approximately 4 ns for Syto-13 dye, approximately 11 ns for the QD585-anti-CD44 ab, and approximately 19 ns for the QD655-anti-EpCAM ab). Lorentzian fitting was used to resolve each spectrum into three with photocount maxima of ~515 nm (Syto-13 nucleus stain), 585 nm (QD585-anti-CD44 ab), and 655 nm (QD655-anti-EpCAM ab). Therefore, by combining three-color imaging with spectrotemporal resolution (the three fluorescence spectral maxima and three fluorescence lifetime values), this experiment showed a 9-fold increase in cancer cell detection accuracy.
The EMT transition causes MCF7 cells to acquire stem-like properties, such as overexpression of the stem cell marker CD4442. The spectral fingerprints and three fluorescence decay data helped us determine the EpCAM and CD44 ratios in this cell line. Considering the comparable fluorescence quantum efficiencies of QD585 and QD655 (approximately 0.8) and using equimolar QD-labeled abs, we found that the expression of the CD44 antigen in MCF7 cells was 4-fold greater than that of EpCAM. This difference was identified by comparing the integrated photocounts of QD585-anti-CD44 ab (20488 photons) and QD655-anti-EpCAM ab (5367 photons) (Fig. 1B). The higher photocount at ~585 nm suggested that CD44 is better than EpCAM for capturing MCF7 cells in a blood sample. Nevertheless, if the energy transfer from QD585 to QD655 is significant, the number of photons counted at ~585 nm (QD585-anti-CD44 ab) is less than the true number. In this case, the expression level of CD44 estimated is more accurate than the estimated value.
Capturing and identifying MCF7 cells using silica particles
We prepared immunosilica microparticles to collect and characterize cancer cells in a blood sample. In addition, we prepared anti-CD44 ab and anti-EGFR ab by biotinylation and stabilized them in PBS. The anti-CD44 ab was tethered to streptavidin-conjugated silica microparticles and purified by repeated washing and gravity separation. The immunosilica particle preparation and cancer cell collection procedures are shown in Fig. 2A, and the related details are presented in the experimental section. Cancer cells from the PBMC or T cells in a blood sample were isolated by gravity separation facilitated by the large size (75 ~ 150 µm) and high specific gravity (~2.2 g/ml) of the silica particles. The sedimentation coefficient of the silica is significantly higher than that of cells due to the difference (approximately. 1.2 kg m−3) in the specific gravities of PBS and silica, allowing gravity separation of the particles in seconds.
A A scheme showing silica particle functionalization and cancer cell separation from a blood sample. B Fluorescence image of silica particles with MCF7 cells. C A Streak camera image and the corresponding fluorescence spectra of cells attached to silica particles. The broken lines represent the Lorentzian fits for the Syto-13 dye (515 nm), QD-CD44-ab conjugate (585 nm), and QD-EpCAM-ab conjugate (655 nm). D, E The fluorescence decays based on the deconvoluted data shown in C for the three labels.
Furthermore, the use of large silica particles helped us prevent endocytosis of silica and magnetic nanoparticles by cells, which is common to traditional CTC isolation. In addition to conjugating anti-CD44-ab to silica particles, we treated a blood sample with a QD585-CD44-ab-blocking solution. Here, the low concentration (6 nM) of QD-ab-labeled conjugate led to the identification of only a portion of CD44 antigens on MCF7 cells, leaving a large portion of the antigens unblocked for cell capture using anti-CD44-ab-conjugated silica particles. We confirmed the presence of excess CD44 antigen by performing a control experiment in which MCF7 cells were labeled with two different concentrations (6 nM and 30 nM) of QD585-anti-CD44-ab solution. After washing the cells, we determined the fluorescence photocounts from an equal number of cells in the samples labeled with 6 nM or 30 nM QD-ab conjugates. The integrated photocounts determined from the streak camera images showed that <30% of CD44 antigens were labeled with a 6 nM conjugate solution. The corresponding fluorescence spectra of different parts of each sample are shown in Fig. S3. The photocount ratios suggest that 60% or more of the CD44 antigens remained intact for potential binding to silica particles.
In the aforementioned method, a mixture of PBMCs and MCF7 cells was prelabeled with the nuclear staining dyes Syto-13, QD585-anti-CD44 ab, and QD655-anti-EpCAM ab. The stained cells suspended in PBS were mixed with anti-CD44-ab-conjugated silica particles, and the MCF7 cells were isolated by gentle mixing of the sample. The silica particles were allowed to settle under gravity in a “natural gravity-mediated enrichment” method. The cancer cells attached to the silica particles were characterized by microspectroscopy. Figure 2B shows a fluorescence image of MCF7 cells attached to anti-CD44-ab-conjugated silica particles, and the corresponding single-photon histogram is shown in Fig. 2C. We deconvoluted the histogram data to obtain a triply degerate fluorescence spectrum (Fig. 2C), and three decay curves (Fig. 2D, E). The characteristic fluorescence spectral maxima (515, 585, and 655 nm) correspond to stained nuclei (Syto-13 dye), CD44 (the QD585-anti-CD44 ab conjugate), and EpCAM (the QD655-anti-EpCAM ab conjugate). Additionally, the decay profiles with the characteristic fluorescence lifetimes of Syto-13, QD585, and QD655 confirmed the attachment of MCF7 cells to the silica particles. The detection of 1–10 cells from 100 MCF7 cells in a 1 mL blood sample was achieved, with the PBMC count consistently maintained at 105. Interestingly, by using anti-CD44 ab to collect MCF7 cells, we avoided the nonspecific adsorption of PBMCs onto silica particles. Only a few PBMCs were nonspecifically attached, as indicated by only the green emission of Syto-13 detected. Thus, we distinguished nonspecifically attached PBMCs from MCF7 cells based on the lack of orange and red fluorescence in the fluorescence images and streak camera histograms. In contrast, multicolor imaging supplemented by spectral and lifetime data helped us identify MCF7 cells.
Estimation of CD44 and EpCAM antigen levels in HeLa cells
To confirm that EMT antigen-specific cancer cells were collected and characterized, we chose HeLa cells in which the expression of epithelial (EpCAM) antigens was downregulated44 and that of EMT-specific CD44 was upregulated. Similar to the labeling procedure used with MCF7 cells, HeLa cells cultured on a tissue culture plate or in a blood sample with the Syto-13 dye, QD585-anti-CD44 ab, and QD655-anti-EpCAM ab were labeled. Figure 3 shows the fluorescence image (3 A) and the temporally and spectrally resolved photocounts (3B) of HeLa cells. In this experiment, we did not detect QD655-EpCAM-ab fluorescence, indicating the poor expression of EpCAM by HeLa cells. Figure 3D shows the fluorescence image of HeLa cells attached to the anti-CD44-ab-conjugated silica particles. First, we treated HeLa cells and PBMCs in a blood sample with Syto-13 dye, a QD585-anti-CD44-ab conjugate, and a QD655-anti-EpCAM-ab conjugate. Then, anti-CD44-ab-functionalized silica particles were added to the blood sample, and HeLa cells were selectively attached to the silica particles by gentle up and down mixing of the sample. The bimodal fluorescence spectrum (Fig. 3C) of the silica particles showing the spectral characteristics of Syto-13 and QD585 and the corresponding fluorescence lifetimes (Fig. S4) confirmed the selective capture of HeLa cells.
A, D Fluorescence images identified with the emission from Syto-13 dye (green) and QD585-anti-CD44 ab (orange) in HeLa cells (A) in a tissue culture dish and D attached to the anti-CD44-ab-conjugated silica particles. B, E Single-photon histograms showing the time- and energy-dispersed photocounts from HeLa cells (A) in a tissue culture dish and D attached to the anti-CD44-ab-conjugated silica particles. C The fluorescence spectrum based on the deconvoluted data shown in E.
The advantage of CD44-targeted CTC collection
To increase the modality of cancer cell detection and minimize false-positive results due to nonspecific adsorption of PBMCs onto silica particles, we used silica particles modified with an anti-EGFR ab. We treated HeLa cells and PBMCs in a blood sample with Syto-25 dye, a QD655-anti-EpCAM-ab conjugate, and an AlexaFluor488-anti-CD44-ab conjugate. In this experiment, the anti-EGFR ab was chosen because EGFR is overexpressed in various cancer cells, indicating that is an alternative biomarker for non-EpCAM-enriched CTCs. Figure 4A shows the fluorescence image of HeLa cells attached to the anti-EGFR-ab-conjugated silica particles. The green-only fluorescence spots in Fig. 4A represent PBMCs attached to silica particles. Undesired binding of PBMCs to anti-EGFR-ab-silica particles is not surprising because of the overexpression of EGFR on PBMCs and T cells12. Figure S5 shows the corresponding single-photon histogram based on the fluorescence spectrum and decay curve characteristic of Syto-25, confirming the capture of PBMCs. In contrast, anti-CD44-ab-conjugated silica particles were ideal for the selective isolation and detection of MCF7 (Fig. 2) and HeLa (Fig. 3) cells.
A A fluorescence image of HeLa cells identified with Syto-25 dye and the AlexaFluor488-anti-CD44-ab conjugate and attached to anti-EGFR-ab-conjugated silica particles. B–D Fluorescence images of anti-EGFR-ab-functionalized silica particles treated in a 1 mL blood sample containing (B) 104 H1650 and 106 T cells, C 104 H1650 cells with no T cells, and D 106 T cells but no H1650 cells.
To verify the collection and detection of cancer cells using anti-EGFR-ab, we chose H1650 cells that upregulate EGFR45. We labeled H1650 and T cells (model blood cells) with Syto-25 dye alone and treated the cell samples with anti-EGFR-ab-conjugated silica particles. Figure 4B–D presents the immunofluorescence images of H1650 cells on silica particles collected from samples with different H1650 and T cell ratios. Figure 4D, which shows a sample with 106 T cells/mL but no H1650 cells, indicates many T cells bound to the immunosilica microparticles. Although EGFR overexpression in many cancer cells46 promises anti-EGFR-ab for their collection, blood cells expressing EGFR interfere with the process. Therefore, the anti-CD44 ab is more promising than the anti-EGFR ab. In contrast, stem cell biomarkers such as CD44 and multiple fluorescence labeling are error-free detection methods for CTC collection and detection. The study presented demonstrates the technical benefits of using mesenchymal biomarkers, spectrally and temporally resolved modalities, and the self-segregating immunosilica particles for CTC detection.
Conclusions
We demonstrated a microspectroscopy method combined with mesenchymal cell-selective immunosilica microparticles for error-free cancer cell detection in which 1–10 cancer cells were accurately identified among 100 cancer cells in a 1 mL blood sample. In this study, we explored cancer cells’ properties for selective attachment to silica microparticles using anti-CD44, anti-EpCAM, or anti-EGFR ab. The large silica particles prevented endocytosis by cells and favored cancer cell isolation without the application of an external centrifugal force. Successively, the bright and narrowband emission of quantum dots helped multiplexed cancer cells detection, which was improved by combining the fluorescence spectra and decay data of the quantum dots and nucleus-staining Syto dye. Furthermore, this research demonstrates the significance of mesenchymal antigen-selective cancer cell collection and characterization. The multicolor imaging and spectrotemporally resolved multimodal fluorescence detection can be further developed for advanced cancer screening and clinical follow-up technologies.
References
Delorey, T. M. et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595, 107–113 (2021).
Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585 (2021).
Kumar, P., Sobhanan, J., Takano, Y. & Biju, V. Molecular recognition in the infection, replication, and transmission of COVID-19-causing SARS-CoV-2: an emerging interface of infectious disease, biological chemistry, and nanoscience. Npg. Asia Mater. 13, 14 (2021).
Keller, L. & Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 19, 553–567 (2019).
Xu, C. & Pu, K. Second near-infrared photothermal materials for combinational nanotheranostics. Chem. Soc. Rev. 50, 1111–1137 (2021).
Shen, Z. et al. Strategies to improve photodynamic therapy efficacy by relieving the tumor hypoxia environment. NPG Asia Mater. 13, 39 (2021).
Laxman, K. et al., Bioinspired carrier-free peptide conjugated BF2-oxasmaragdyrin dye-based nano self-assemblies: a photostable NIR cancer theragnostic agent. NPG Asia Mater. 12, 75 https://doi.org/10.1038/s41427-020-00256-x (2020).
Hodgkinson, C. L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Alix-Panabieres, C. & Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–631 (2014).
Shen, Z., Wu, A. & Chen, X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 46, 2038–2056 (2017).
Li, W. et al. Emerging nanotechnologies for liquid biopsy: the detection of circulating tumor cells and extracellular vesicles. Adv. Mater. 31, 1805344 (2019).
Baccelli, I. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31, 539–544 (2013).
Ashworth, T. R. A Case of Cancer in Which Cells Similar to Those in the Tumours Were Seen in the Blood after Death. T. R. Med. J. Aust. 14, 146 (1869).
Kim, M. Y. et al. Tumor self-seeding by circulating cancer cells. Cell 139, 1315–26 (2009).
Ring, A., Smith, I. E. & Dowsett, M. Circulating tumour cells in breast cancer. Lancet Oncol. 5, 79–88 (2004).
Cabel, L. et al. Clinical utility of circulating tumour cell-based monitoring of late-line chemotherapy for metastatic breast cancer: the randomised CirCe01 trial. Br. J. Cancer 124, 1207–1213 (2021).
Bidard, F. C. et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 15, 406–414 (2014).
de bono, J. S. et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14, 6302–6309 (2008).
Khoo, B. L. et al. Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci. Adv. 2, e1600274 (2016).
Nagrath, S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450, 1235–1239 (2007).
Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med 351, 781–791 (2004).
Wu, L. L. et al. Spectrally combined encoding for profiling heterogeneous circulating tumor cells using a multifunctional nanosphere‐mediated microfluidic platform. Angew. Chem. Int. Ed. 59, 11240–11244 (2020).
Schiro, P. G. et al. Sensitive and high-throughput isolation of rare cells from peripheral blood with ensemble-decision aliquot ranking. Angew. Chem. Int. Ed. 51, 4618–4622 (2012).
Yoon, H. J. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 8, 735–741 (2013).
Wang, S. et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew. Chem. Int. Ed. 50, 3084–3088 (2011).
Park, M. H. et al. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip. J. Am. Chem. Soc. 139, 2741–2749 (2017).
Poudineh, M. et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 12, 274–281 (2017).
Lu, N. N. et al. Biotin-triggered decomposable immunomagnetic beads for capture and release of circulating tumor cells. ACS Appl. Mater. Interfaces 7, 8817–8826 (2015).
Rosenholm, J. M. et al. Targeting of porous hybrid silica nanoparticles to cancer cells. ACS Nano 3, 197–206 (2009).
Wang, L., Asghar, W., Demirci, U. & Wan, Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today 8, 374–387 (2013).
Yoo, C. E. et al. Highly dense, optically inactive silica microbeads for the isolation and identification of circulating tumor cells. Biomaterials 75, 271–278 (2016).
Biju, V., Itoh, T. & Ishikawa, M. Delivering quantum dots to cells: bioconjugated quantum dots for targeted and nonspecific extracellular and intracellular imaging. Chem. Soc. Rev. 39, 3031–3056 (2010).
Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 43, 744–764 (2014).
Anas, A. et al. Clathrin-mediated endocytosis of quantum dot-peptide conjugates in living cells. ACS nano 3, 2419–2429 (2009).
Shibu, E. S. et al. Photouncaging nanoparticles for MRI and fluorescence imaging in vitro and in vivo. ACS Nano 7, 9851–9859 (2013).
Payne, R. E. et al. Measurements of EGFR expression on circulating tumor cells are reproducible over time in metastatic breast cancer patients. Pharmacogenomics 10, 51–57 (2009).
Flores, L. M. et al. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. Br. J. Cancer 102, 1495–502 (2010).
Lecharpentier, A. et al. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br. J. Cancer 105, 1338–1341 (2011).
Yu, M. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013).
Mohamadi, R. M. et al. Nanoparticle‐mediated binning and profiling of heterogeneous circulating tumor cell subpopulations. Angew. Chem. Int. Ed. 54, 139–143 (2015).
Grillet, F. et al. Circulating tumour cells from patients with colorectal cancer have cancer stem cell hallmarks in ex vivo culture. Gut 66, 1802–1810 (2017).
Chen, C., Zhao, S., Karnad, A. & Freeman, J. W. Anti-GD2/4-1BB chimeric antigen receptor T cell therapy for the treatment of Chinese melanoma patients. J. Hematol. Oncol. 11, 1 (2018).
Poortmans, P. M. et al. Internal mammary and medial supraclavicular irradiation in breast cancer. N. Engl. J. Med. 373, 317–327 (2015).
Cheng, B. et al. Quantification of rare cancer cells in patients with gastrointestinal cancer by nanostructured substrate. Transl. Oncol. 7, 720–725 (2014).
Shibu, E. S. et al. Singlet-oxygen-sensitizing near-infrared-fluorescent multimodal nanoparticles. Angew. Chem. Int. Ed. 52, 10559–10563 (2013).
Kawashima, N. et al. Reversible dimerization of EGFR revealed by single-molecule fluorescence imaging using quantum dots. Chem. Eur. J. 16, 1186–1192 (2010).
Acknowledgements
V.B. acknowledges a MEXT, JSPS grant-in-aid for scientific research (grant # 19H02550), the Hokkaido University Photoexcitonix Program, and the JSPS Dynamic Alliance for Open Innovation Bridging Human, Environment, and Materials. J.S. thank the Special Grant Program of the Graduate School of Environmental Science, Hokkaido University.
Author information
Authors and Affiliations
Contributions
V.B. conceived the idea and led the project. J.S. conducted chemical and bioconjugate reaction experiments and cell imaging. J.S. and S.S. labeled cells. E.H. collected blood samples. S.Y. and Y.T. provided blood cells. J.S. and V.B. identified cancer cells on silica particles. J.S. wrote the manuscript draft. J.S., Y.T., and V.B. finalized the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics
We collected and used PBMCs obtained from a healthy adult donor following the procedure approved by the Ethics Committee (No. 2020–7) at Hokkaido University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sobhanan, J., Takano, Y., Sugino, S. et al. Multimodal CTC detection using stem cell antigen-specific immunosilica particles and immunofluorescent quantum dots. NPG Asia Mater 14, 3 (2022). https://doi.org/10.1038/s41427-021-00353-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41427-021-00353-5