Abstract
Recent advances in three-dimensional (3D) cell culture models developed on organ-on-a-chip or microfluidic devices have shown their capability to recapitulate the in vivo microenvironment as well as their potential as tools in biomedical research. Here, we present an in vitro model of the peripheral nervous system (PNS) by establishing a coculture model of motor neurons (MNs) and Schwann cells (SCs) in a 3D environment in a microengineered extracellular matrix hydrogel scaffold. The collagen scaffold placed at the center of the microdevice provided a 3D cellular microenvironment where the axons of MNs were allowed to actively interact with SCs during their growth and maturation. By treating the MN–SC coculture model with ascorbic acid, we were able to model the myelination process in the PNS, which was evidenced by the increased expression of myelin markers in SCs. Moreover, we show that this can be reversed by treating myelinated nerve fibers with glial growth factor (neuregulin-1 isoform) to potentially block the formation of the myelin sheath and induce demyelination. Our 3D cell culture model may be used to achieve active control of the myelinating and demyelinating processes in the PNS and thus may offer new opportunities to study pathophysiological processes involved in motor neuron diseases by in vitro modeling.
Similar content being viewed by others
Introduction
In the peripheral nervous system (PNS), the continuous bidirectional interaction between axons and Schwann cells (SCs) is important for myelin formation1,2. Axonal signals are responsible for SC migration, proliferation, and differentiation into myelinating or nonmyelinating cells3. Reciprocally, SCs provide trophic support for neuronal maturation, and their signaling induces the sequential assembly of proteins in the process of myelination to organize domains that are essential for efficient action potential propagation4. Although the characteristics and potentialities of SCs have been identified, the ability to study axon–SC interactions in a three-dimensional (3D) environment would advance our understanding of physiological interactions between motor neurons (MNs) and SCs during myelination and demyelination processes.
To better recapitulate physiologically relevant models, it is important that such model systems move beyond traditional cell cultures to models that specifically mimic the physiological and biochemical aspects of organs or tissues. A variety of in vitro myelination models of PNS have been developed to study demyelinating diseases, where pathological processes are expected to mimic human diseases (Supplementary Table 1)4,5. However, they do not allow accurate replication of the PNS since traditional in vitro models are mainly developed in two-dimensional (2D) cell culture settings where neurons are cultured on the surface of a coverslip with a random orientation6,7. To create a physiologically relevant environment, it is important that such a model system recapitulate the 3D aspects of specific organs or tissues. More specifically, in our body, the cell bodies and dendrites of MNs are located in the spinal cord (CNS) to receive signals, whereas SCs and the axons of MNs are located in the PNS and form myelinated nerve fibers to deliver signals to the target muscle (Fig. 1a). Recently, various microfabricated platforms have been developed to enable the coculture of spatially regulated cells at the cellular microenvironment scale8,9. By taking advantage of microfabrication technology, it is possible to establish a PNS myelination model in a 3D microenvironment that would improve our ability to study motor neuron diseases in a more physiologically relevant context.
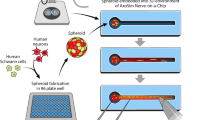
a In the peripheral nervous system, the axons of motor neurons (MNs) closely interact with Schwann cells (SCs) to transmit signals to the target tissues. b Isolation of MNs and the MN–SC coculture procedure. MNs were isolated from the mouse lumbar spinal cord on embryonic day 13 (E13). Dissected spinal cords (L1-L6) were trypsinized and incubated in a p75 panning plate for 45 min to allow the isolation of MNs from other types of cells in the spinal cord. After incubation, a depolarization solution was applied to detach the MNs, and the isolated cells were seeded throughout the medium channel. c Image of the microengineered device used in this study for the 3D MN–SC coculture. d The platform consists of two medium channels on the sides and one extracellular matrix (ECM) hydrogel channel in the middle, which are separated by PDMS posts (height: 200 µm). MNs are suspended through the medium channel and then allowed to attach onto the ECM hydrogel. SCs are restricted inside the ECM hydrogel by encapsulation in Col I hydrogel. The hydrogel channels in the 3D microfluidic cell culture platform isolate SCs and axons from the soma of MNs. e MNs attached to the surface of the ECM hydrogel show spherical formation on day 1. MNs were identified by NeuN (red) and DAPI (blue) staining. f To confirm axonal outgrowth, dendrites were identified by immunostaining for Map2 (red), and axons were identified by immunostaining for Tau (green). Nuclei were identified with DAPI (blue) on day 3 of culture. g Confocal image illustrating axonal growth by immunostaining for TuJ1 (red), demonstrating that neurites reached the end of the hydrogel channel on day 5 of MN monoculture. Scale bars: (d) 500 μm, (e, f) 50 μm, (g) 100 μm.
Here, we demonstrate the use of a microfabrication technique to develop a long-term cell culture platform that can be used for 3D compartmentalization of primary MNs and SCs. We describe the fabrication and assessment of a feasible 3D cell culture platform in which the SCs and MN axons are located within the extracellular matrix (ECM) hydrogel and the polarity of axonal outgrowth can be achieved with spatial separation from the MN cell bodies and dendrites. Using this microengineered ECM scaffold, we investigated the cellular behaviors of MNs and SCs in a 3D environment. Then, the effect of the MN–SC coculture on cell growth and maturation was studied. Our model was further developed into a peripheral myelination model by treating it with ascorbic acid (AA), which is known to regulate basal lamina and myelin formation by SCs by increasing the expression of myelin-related genes10,11. To demonstrate the potential application in disease modeling, myelinated nerve fibers were treated with a high concentration of glial growth factor (GGF, neuregulin-1 isoform) to block myelination and to produce a demyelination model12. Taken together, our results indicate that our 3D in vitro PNS model has utility for the assessment of PNS myelination and demyelination by culturing MNs and SCs within the microengineered device. The model established in this study may have significant applications for motor neuron diseases by providing a drug screening platform for developing novel therapeutics.
Materials and methods
Fabrication of the silicon wafer and PDMS chip
The 3D microfluidic platform was fabricated based on the protocol described in our previous study8. Briefly, a cleaned and dehydrated 4-inch silicon wafer was spin-coated with SU-8 resist at a thickness of 200 µm with patterning and prebaked on a hot plate at 65 °C for 20 min and at 95 °C for 50 min (Supplementary Fig. 1). The coated wafer was exposed to UV light and postbaked on a hot plate at 65 °C for 1 min and at 95 °C for 12 min. The wafer was then developed in a photoresist developer for 15 min and finally rinsed with isopropyl alcohol (IPA) and dried with pressurized nitrogen gas. Polydimethylsiloxane (PDMS) devices were fabricated using soft lithography. Sylgard 184 silicone elastomer base (Dow Corning, USA) and the curing agent were combined at a ratio of 10:1 (w/w), mixed thoroughly, and poured over the master wafer at a thickness of 5 mm. Air bubbles were removed by degassing the wafer under a vacuum, and the PDMS was then allowed to polymerize in the oven for 24 h at 80 °C. The cured PDMS was removed from the wafer using a scalpel, and the holes through the reservoir patterns were created using a 4 mm disposable biopsy punch. For the hydrogel filling inlets, a 1 mm disposable biopsy punch was used. Small particles on the PDMS were removed using Scotch tape, and the PDMS was placed in a glass beaker filled with deionized (DI) water and sterilized using an autoclave. The devices were dry autoclaved once more by placing the PDMS in a clean box with the channel sides facing up. The sterilized devices were incubated in the oven for 24 h at 80 °C. Devices were then placed in plasma cleaner with glass coverslips, and the treated PDMS surface was bonded to the glass coverslip using sterile tweezers. The bonded devices were placed in the oven for 24 h at 80 °C and stored at room temperature until use.
Collagen gel preparation and filling procedure
To make the collagen gel solution, 20 µL of 10X phosphate buffered saline (PBS) with phenol red was added into a sterile 1.5 mL tube (Supplementary Fig. 2). Then, 5 µL of 0.5 N NaOH was carefully added into the same tube followed by 58.4 µL of DI water. Finally, a total volume of 116.6 µL of collagen type I, rat tail (Col I; BD Bioscience, cat. #40236) stock solution (stock concentration: 3.43 mg/mL) was added and carefully mixed until the gel appeared uniformly distributed. The final concentration of the Col I solution was set to 2 mg/mL, and the pH of the resultant solution was measured (pH 7.4). To fill the PDMS device with gel, a prewarmed humid chamber was prepared. The Col I gel solution was carefully injected into the hydrogel scaffold region using a 10 µL micropipette. After gel filling, the devices were placed in a humid chamber with the channel side facing up. The humid chamber was then placed in a CO2 incubator at 37 °C for 30 min to allow gelation. After incubation, the medium channels of the device were filled with the appropriate cell culture medium, and the device was placed in a CO2 incubator until addition of the cell suspension.
Primary motor neuron culture
Motor neurons (MNs) were isolated from the lumbar spinal cord of CD-1 mice (E13) according to a previously described procedure13. Carefully, the lumbar region of the spinal cord (L1–L6 segment) was collected separately in a sterile 1.5 mL tube with 180 µL of Hank’s balanced salt solution (HBSS) on ice (Fig. 1b). Then, 20 µL of a 1% trypsin solution (Worthington, USA) was added and carefully mixed. Trypsinization was performed by placing the tubes in a 37 °C water bath for 15 min. After incubation, trypsinization was stopped by adding 20 µL of 1% trypsin inhibitor (Sigma, USA), and gentle trituration was performed using a 200 µL pipette tip. Cells were transferred to a prepared panning plate with a p75NTR antibody solution (1:5000; Abcam, UK) covered with GlutaMAX 1-containing (Gibco, USA) neurobasal medium (Gibco, USA). The panning plate was sealed with parafilm and incubated for 45 min at room temperature. The plate was washed three times with prewarmed neurobasal medium containing GlutaMAX 1, then with 300 µL of depolarization solution containing 30 mM potassium chloride and 0.8% sodium chloride; and 2 mM calcium chloride was added after the last washing step to detach the cells from the panning plate. After 10 s, 500 µL of prewarmed MN culture medium composed of neurobasal medium, 10% horse serum (HS; Gibco, USA), 1x GlutaMAX 1, 1x B27 supplement (Gibco, USA), 1 µM β-mercaptoethanol (Sigma, USA), and 10 ng/mL BDNF (Gibco, USA) was added. Cells were collected in a sterile 15 mL tube by pipetting them up and down and then centrifuged at 400g for 5 min at room temperature. After centrifugation, the cells were resuspended in MN culture medium and seeded on the prepared device by adding 60 µL of cell suspension into the reservoir at a density of 1 × 06 cells/mL. The devices were placed in an incubator containing CO2 for 1 h to allow cell attachment, and the medium was replaced by adding 60 µL of fresh MN culture medium to each reservoir.
Schwann cell culture
Mouse SCs were obtained from ScienCell Research Laboratories (#M1700; USA). SCs were maintained in basal medium supplemented with 5% fetal bovine serum (FBS), 1% SC growth supplement (SCGS), and 1% penicillin/streptomycin (P/S) solution.
Motor neuron–Schwann cell coculture
For the MN–SC coculture, mouse SCs were mixed in a Col I solution and seeded into the device through the hydrogel scaffold region at a density of 5 × 105 cells/mL 7 days prior to MN culture. SCs were cultured for 1 week with SC medium composed of basal medium (#1701; ScienCell, USA), 5% FBS (ScienCell, USA), 1% SCGS (ScienCell, USA), and 1% P/S solution. On day 0, MNs were seeded by adding 60 µL of the cell suspension into the reservoir at a density of 1 × 106 cells/mL. Until day 7, MN–SC coculture medium composed of neurobasal medium, 10% HS, 0.5 mM L-glutamine (L-gln; Gibco, USA), 1x B27 supplement, 1 µM β-mercaptoethanol, 0.5 µM forskolin (Sigma, USA), 1 µg/mL bovine pituitary extract (Thermo Fisher Scientific, USA), and 10 ng/mL BDNF was refreshed daily. After day 7, 50 µg/mL ascorbic acid (AA; Sigma, USA) was added to the MN–SC coculture medium to induce myelination. To induce demyelination, 200 ng/mL GGF (neuregulin-1 isoform) was added to the AA-supplemented MN–SC coculture medium starting on day 14.
Enzyme-linked immunosorbent assay
The concentrations of nerve growth factor (NGF) and glial cell-derived neurotrophic factor (GDNF) were measured using mouse NGF (CSB-E04684m; CUSABIO, USA) and GDNF (ab171178; Abcam, UK) enzyme-linked immunosorbent assay (ELISA) kits. Using these kits, the minimum levels of NGF and GDNF detection were 1.56 and 31.2 pg/mL, respectively. In brief, 100 µL of standard or sample was added in duplicate, and the plate was incubated for 2 h at 37 °C. After incubation, the liquid in each well was removed, and 100 µL of biotin antibody (1x) was added to each well. After 1 h of incubation at 37 °C, the plate was washed three times with 200 µL of wash buffer, and 100 µL of HRP-avidin (1x) was added to each well and incubated for 1 h at 37 °C. After washing 5 times, 90 µL of TMB substrate was added to each well and incubated for 25 min at 37 °C protected from light. Then, 50 µL of stop solution was added, and the optical density of NGF or GDNF was detected immediately at a 450 nm wavelength. The final concentration of NGF or GDNF was calculated according to the fitted standard curve.
Antibodies
The following antibodies were used in this study: mouse monoclonal anti-β3-tubulin (TuJ1; ab78078, 1:1000), rabbit polyclonal anti-MAP2 (ab32454, 1:1000), rabbit polyclonal anti-β3-tubulin (TuJ1; ab18207, 1:1000), mouse monoclonal anti-200 kD-neurofilament heavy (NF-H; ab7795, 1:1000), rabbit polyclonal anti-200 kD-neurofilament heavy (NF-H; ab8135, 1:1000), rabbit polyclonal anti-myelin basic protein (MBP; ab40390, 1:200), rabbit polyclonal anti-PMP22 (ab211052, 1:200), rabbit polyclonal anti-myelin protein zero (P0; ab31851, 1:100), rabbit polyclonal anti-Caspr (ab34151, 1:1000), rabbit monoclonal anti-S100β (ab52642, 1:100), mouse monoclonal anti-Tau (ab80576, 1:200), mouse monoclonal anti-160 kD-neurofilament medium (NF-M; ab7794, 1:1000), goat anti-rabbit IgG H&L (TRITC; ab6718, 1:1000), goat anti-mouse IgG H&L (FITC; ab6785, 1:1000), and CytoPainter Phalloidin-iFluor 488 reagent (ab176753, 1:1000) from Abcam (UK).
Immunofluorescence assay
Cells cultured on microfluidic devices or coverslips were rinsed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature. The fixed samples were washed twice with PBS, and 60 µL of 0.1% Triton X-100 (Thermo Fisher Scientific, USA) was added to each reservoir for 3 min at room temperature. Then, the devices were washed twice with PBS, and 60 µL of 3% BSA (Sigma, USA) was added and incubated for 24 h at 4 °C. After washing the devices with PBS, the samples were incubated for 24 h at 4 °C with the primary antibodies. The devices were then washed twice with PBS and incubated with the secondary antibodies for 24 h at 4 °C. For nuclear staining, DAPI (D1306, 1:1000), which was obtained from Life Technologies (USA), was added to the secondary antibody solution. After washing the devices twice with PBS, all reservoirs were filled with 60 µL of fresh PBS and placed on a shaker for 24 h at 4 °C. Confocal fluorescence images were captured on a confocal laser scanning microscope (LSM 700; Carl Zeiss, Germany) using a 20X/0.8 NA objective. Images were acquired by creating confocal z-stacks spanning from the minimum to the maximum depth of the ECM hydrogel region with 1 µm intervals, each of which had a resolution of 1024 × 1024. The obtained images were processed using ZEN 2011 (Zeiss, Germany) and ImageJ (National Institutes of Health, USA) with the projections of the z-stacks at the maximum intensity.
Transmission electron microscopy
Samples were fixed with 2.5% glutaraldehyde (Sigma, USA) in 0.1 M PBS (pH 7.4) for 24 h at 4 °C. After washing with 0.1 M PBS, the samples were postfixed with 1% osmium tetroxide (OsO4) in 0.1 M sodium cacodylate trihydrate buffer for 1 h at room temperature. Then, the samples were dehydrated in a series of ethanol solutions (70%-80%-85%-90%-95%-100%-100%) followed by dehydration twice in pure propylene. Samples were embedded with the Epoxy Embedding Medium Kit (Sigma, USA). The silicone membranes were detached from the PDMS chip, and the embedded samples were removed from the microchannels. The samples were then cut into ultrathin sections (70–80 nm) using an ultramicrotome (Leica, Germany) and stained with uranyl acetate and lead citrate for transmission electron microscopy (TEM) imaging. Cryo-TEM (CryoTecnai F20; FEI, USA) was used to observe the samples.
Statistical analysis
For the statistical analysis, one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was performed for the between-group comparisons, and the following P values were considered statistically significant: *P < 0.05, **P < 0.01, and ***P < 0.001. All data are expressed as the mean ± standard error of the mean (SEM).
Results and discussion
Establishment of an in vitro peripheral nervous system model in a three-dimensional microenvironment
Our microengineered cell culture platform consists of two medium channels on the side and a hydrogel channel in the middle (Fig. 1c). The design of the device allows SCs to be encapsulated in a Col I hydrogel separately from the cell bodies of MNs (Fig. 1d). In addition, the Col I gel in the hydrogel channel allows the axon but not the cell body to grow into the ECM hydrogel. It is important to note that this is an important feature of PNS myelination, and by mimicking this architecture, our device allows focused investigation of axon–SC interactions during the myelination process.
Prior to investigating the characteristics of MNs in our device, we used the neuron-specific marker NeuN to confirm the identify of MNs isolated from spinal cords (Fig. 1e). In addition, MAP2 (dendrites) and tau (axons) antibodies were used to measure the length of the neurites and evaluate dendritic and axonal outgrowth in MN monocultures. On day 1 of MN culture, the axons began to grow in the ECM hydrogel. On day 3 of the culture, the axons were observed to extend significantly into the ECM hydrogel, while the dendrites largely remained close to the cell bodies of MNs, as assessed by immunocytochemistry (Fig. 1f). On day 5, the neurites extended by 1300 µm and reached the end of the hydrogel channel (Fig. 1g). Collectively, these data suggest that primary MNs can be cultured in our device, where axons are able to grow longer than 1 mm in a 3D microenvironment.
The role of SCs in axonal outgrowth
Next, we examined axonal outgrowth in the 3D microenvironment with and without SCs. Interestingly, significantly more axonal outgrowth was observed from day 1 of the MN monoculture compared to dendritic outgrowth (112.0 ± 16.0 and 46.6 ± 5.2 µm, respectively) (Fig. 2a, b). This tendency continued on days 3 and 5, as shown by the lengthening of axons (252.9 ± 27.0 and 569.2 ± 41.3 µm, respectively), whereas the majority of dendrites remained near the cell body of the MNs (65.6 ± 4.1 and 111.9 ± 7.8 µm, respectively) (Fig. 2a, b).
a Axons (Tau, green) of MNs were isolated from somas (DAPI, blue) or dendrites (MAP2, red) by the ECM hydrogel. Axons extended from the PDMS posts and grew into the hydrogel channel. b Quantification of the axonal and dendritic lengths of MNs (n = 6, mean ± SEM; **P < 0.01 and ***P < 0.001 according to a Tukey post hoc comparison after one-way ANOVA). c Neurites were immunostained with the neuronal marker TuJ1 on days 1, 3, and 5 of MN monoculture or MN–SC coculture. Scale bars, 50 µm. d Quantification of the extended neurite length inside the ECM hydrogel channel shows the faster outgrowth of neurites in the MN–SC coculture (n = 6, mean ± SEM; ***P < 0.001, Tukey post hoc comparison after one-way ANOVA). Scale bars: (a, c) 50 μm.
We then investigated the role of SCs in axonal outgrowth by comparing the MN monoculture and MN–SC coculture. Significant axonal outgrowth was observed starting on day 1 in the SC coculture, and axons reached the end of the hydrogel channel on day 3 (Fig. 2c). However, the relative axonal growth rate of the MN monoculture was slower than that of the MN–SC coculture. The mean neurite length on day 1 of the MN–SC coculture was 145 ± 16.5 µm, which was approximately three times longer than that of the MN monoculture (Fig. 2d). This difference was also observed on day 3 of culture, but the neurites reached the maximum length (width of the hydrogel channel, 1300 µm) by day 5 in both the MN–SC coculture and MN monoculture. These results support the role of SC in axonal outgrowth via the secretion of NGFs, which are crucial for establishing axonal regeneration and PNS regrowth following nerve injury14. Previous studies of cells cultured on a 2D coverslip have reported that the axonal length in MN monocultures and MN–SC cocultures on day 2 was less than 50 and 150 µm, respectively, with an axonal growth rate of ~50 µm/day7. Thus, it is important to note that for MNs cultured in our platform, the axons grow five times faster than axons in cells cultured on 2D surfaces. Together, the results reveal that the 3D microenvironment and the presence of SCs accelerate axonal growth. However, it should be noted that the underlying mechanism of the chemical interactions between axons and SCs should be further investigated to provide a clear understanding of the role of SCs in axonal growth.
Morphological phenotype and alignment of Schwann cells in a 3D microenvironment
In vivo, SCs need to maintain their bipolar phenotypes to function in a biomechanical environment. However, when cultured in vitro, they exhibit motile phenotypes with three different forms: unpolarized, bipolar, and multipolar15,16. We compared SCs cultured in the ECM hydrogel to those in 2D cultures to investigate the role of the 3D microenvironment in the phenotypic expression of SCs in the absence of MNs. First, mouse SCs were cultured on the microfluidic platform (3D) or a poly-D-lysine (PDL)-coated coverslip (2D) for 7 days (Fig. 3a). In both the 3D and 2D cultures, SCs displayed three morphological phenotypes when actin filament (F-actin) expression was visualized. The unpolarized phenotype showed a circular shape, with the nucleus located at the center of the SC. The bipolar and multipolar phenotypes were characterized by extensions (two for bipolar cells and three or more for multipolar cells) far from the nucleus. The different morphological phenotypes of SCs can undergo spontaneous transformation to different cell motility phenotypes during morphogenesis. To compare the percentages of the different cell motility phenotypes between the 3D and 2D in vitro cultures, the SC cytoskeleton was visualized with an F-actin antibody on days 1, 3, 5, and 7, and the number of cells displaying each phenotype was measured. The effect of the spatial environment on the SC phenotype was assessed by the percentage occurrence of each morphological phenotype in the 2D and 3D cell cultures (Fig. 3b). Three different morphological phenotypes of SCs were observed in vitro: unpolarized, bipolar, and multipolar. On the first day of 2D culture, SCs mainly displayed an unpolarized phenotype (60.7%), with 17.9% of cells displaying a bipolar phenotype. These percentages remained similar on day 7, with 48.4% of cells showing an unpolarized phenotype and 25.8% showing a bipolar phenotype. The percentage of each morphological phenotype in the 3D culture was similar to that in the 2D culture on day 1, with 52% of cells showing an unpolarized phenotype and 20% showing a bipolar phenotype. However, on day 7, 56.5% of SCs displayed a bipolar phenotype, and 10.5% showed an unpolarized phenotype. Because the bipolar phenotype is known to be an important feature of motile and migratory SCs in the PNS15, our findings demonstrate that the Col I ECM-based scaffold created in our device played a critical role in providing a 3D environment that allows the spontaneous transformation of SCs into the bipolar phenotype, which is similar to that observed in vivo17.
a Confocal micrographs of 2D- and 3D-cultured SCs on day 3 showing three different morphological phenotypes. b Quantification of temporal changes in the number of SCs expressing each morphological phenotype (unpolarized, bipolar, and multipolar) (n = 5). c, d SCs were cultured in a microfluidic platform in the absence of axons, and images of tracings were acquired on days 3 and 7 for S100β+ cells. e, f SCs were cocultured with MNs to demonstrate cell alignment prior to myelination. SCs were immunostained for S100β (red) and neurites were immunostained for TuJ1 (green). From the confocal images, SC and axon tracings were acquired using NeuronJ. g–i From the SC and axon tracings, the orientation of cells was measured in degrees and is represented as the frequency distribution for the (g) SC monoculture and (h, i) MN–SC coculture (n = 30). j Confocal images of SCs aligning with nearby axons. Neurites were immunolabeled with TuJ1, and SCs were immunolabeled with S100β. k Temporal change in the frequency distribution of the angles between axons and SCs indicates the presence of highly aligned SCs on day 7 of coculture (n = 30). Scale bars: (a) 50 μm, (c–f) 100 μm, (j) 50 μm.
We then examined the effect of axon–glial signaling on the morphological orientation of SCs. For the analysis, we immunostained SCs and MNs with S100β and TuJ1 antibodies, respectively, and obtained their tracings using ImageJ (Fig. 3c–f). SCs were aligned with the proximal axons on day 7 of coculture; however, a random distribution of SC orientations was observed when SCs were cultured without MNs (Fig. 3g–i). Prior to initiating myelination, SCs migrate along outgrowing axons and communicate with them to provide a supportive environment for neurite outgrowth through the release of neurotrophic factors, expression of cell surface ligands, and synthesis of ECM (Fig. 3j)18,19,20. The angle between the SC and the axons was measured to assess the alignment of the SC and adjacent axons (Fig. 3k). SCs cultured in the ECM hydrogel showed a random orientation on day 3 of the MN–SC coculture. However, on day 7, SCs closely associated with axons by aligning in the same direction via a physiological process similar to that which occurs in vivo during radial sorting21,22. The angle (θ) measured between SCs and axons confirmed their alignment on days 5 and 7 of MN–SC coculture by exhibiting a high bias in the frequency distribution for θ < 15°. Our results suggest a tendency for SCs to migrate in the direction of proliferating axons prior to ensheathment, which also indicates that axon–SC interactions are associated with SC-induced directional neurite outgrowth and axonal-induced SC alignment23,24.
Maturation of neurites in the myelination and demyelination models
We then investigated the effect of SCs on neuronal maturation by immunolabeling neurites with the NF-M (160 kD) and NF-H (200 kD) antibodies. During the MN–SC coculture, ascorbic acid (AA) was added starting on day 7 to initiate myelination, and then GGF was added starting on day 14 to induce demyelination (Fig. 4a). In both the MN monoculture and MN–SC coculture, the mature neurites expressed both NF-M and NF-H (Fig. 4b, c). When the numbers of axons expressing NF-M and NF-H were counted, there was a significant difference in the number of NF-M+ neuronal segments on day 21 of culture (Fig. 4d). While there were 100.1 ± 18.4 neurites/mm2 of NF-M+ axons in the MN monoculture, this number was significantly increased to 156.3 ± 9.8 neurites/mm2 of NF-M+ axons in the MN–SC coculture model. In addition, a significant difference in the number of NF-H+ neurites between the MN monoculture and the MN–SC coculture was observed on day 7 (15.6 ± 7.3 and 48.8 ± 6.9 neurites/mm2, respectively) (Fig. 4d). Furthermore, GGF treatment in the MN–SC coculture model significantly reduced the number of NF-H-expressing neurites on day 21. These results indicate that SC plays a critical role in neuronal maturation, as NF-H expression is low in developing neurons and increases in large, myelinated axons, and this tendency can be reversed in a demyelination model by GGF treatment. Furthermore, when treated with AA, the synthesis of collagen type 4 (Col 4) increased in the myelination model (Fig. 4e, f), supporting previous findings that showed Col 4 serves as a scaffold for the components of the basal lamina that attract SCs to attach to axons and initiate myelination10,25.
a MN–SC coculture method in a microfluidic platform. SCs are embedded in the Col 1 ECM inside the hydrogel channel. After 1 week of SC culture, MNs are seeded inside the medium channel. On day 7 of the coculture, ascorbic acid (AA) was added to the MN–SC coculture medium to induce myelination. In addition, glial growth factor (GGF) was supplemented along with AA to develop the demyelination model on day 14 of MN–SC coculture. Immunostaining of axons for NF-M (160 kD) and NF-H (200 kD) in the b MN monoculture and c MN–SC coculture. d Quantification of neuronal maturation by counting the number of NF-M+ and NF-H+ axonal segments on days 7, 14, and 21 (n = 6, mean ± SEM; *P < 0.05 and **P < 0.01, Tukey post hoc comparison after one-way ANOVA). e ECM (Type IV collagen and laminin) deposits by SCs. SCs were cultured alone in the Col I ECM hydrogel with (demyelination model) or without (myelination model) GGF treatment. f Abundant Col 4 expression was observed in the myelination group on day 14 (n = 6, mean±SEM; ***P < 0.001, Tukey’s post hoc comparison after one-way ANOVA). Scale bars: (b, c, e) 50 μm.
Myelination and demyelination models of the PNS
In our device, supplementation of AA in the MN–SC coculture medium induced rapid myelin formation around the axonal segments, which was clearly visible in tubular structures in phase-contrast microscopy images obtained on days 14 and 21 (data not shown). We then further analyzed myelin formation by immunolabeling MBP. To quantify the proportion of SCs involved in the myelination process, MBP+ cells were counted to determine the number of myelinating SCs (Fig. 5a). Terminally differentiated SCs forming compact myelin are known to express MBP26,27, and MBP expression was detected in the MN–SC coculture model on days 14 and 21 (Fig. 5b). On day 14 of the MN–SC coculture, the SCs were mostly in the premyelinating stage, with only 28.7 ± 5.0% of cells expressing MBP (Fig. 5c). However, a higher percentage of SCs expressing MBP was observed on day 21 (75.9 ± 11.3%), indicating that the majority of SCs at this stage were in the myelinating process and had formed fully myelinated axonal segments. In contrast to the myelination model, GGF treatment in the demyelination model reduced the number of myelinating SCs on day 21 (44.2 ± 5.1%), indicating that maturation and myelination were suppressed in the demyelination model.
a Illustration of the ensheathment and wrapping processes of SCs in the premyelinating and myelinating stages. b Images of MBP+ axonal segments in the myelination and demyelination models. Axons are immunolabeled with NF-H. c Quantification of MBP-expressing cells to assess the proportion of myelinating SCs in the myelination and demyelination models (n = 6, mean ± SEM). The concentrations of (d) nerve growth factor (NGF) and (e) glial cell-derived neurotrophic factor (GDNF) in the MN monocultures and MN–SC cocultures were measured by ELISA at different time points. d In the case of the MN–SC coculture, both the myelination and demyelination models showed significantly increased NGF concentrations, whereas the NGF concentration in the MN monoculture remained low (n = 3, mean of three independent experiments; *P < 0.05 and ***P < 0.001, Tukey post hoc comparison after one-way ANOVA). e For GDNF, there was no significant difference between the myelination and demyelination models throughout culture (n = 3, mean of three independent experiments; **P < 0.01, Tukey post hoc comparison after one-way ANOVA). Myelin formation was further confirmed by immunostaining for: f, g PMP22 and h, i P0 to visualize the SC wrapping regions in axonal fibers in the myelination and demyelination models. j High-magnification confocal microscopic images of MBP+/Caspr+ axons revealing the nodes of Ranvier that formed between MBP-expressing segments. Green arrows indicate myelin formation, and red arrows indicate formation of nodes of Ranvier. Scale bars: (b, f, h) 50 μm, (j) 10 μm.
In the nervous system, glial cells are known to provide growth factors that support neuronal development28,29. In our model, we previously found that SC plays an essential role in neuronal outgrowth and maturation. Specifically, when MNs were cultured with SCs, a significant increase in NGF was observed (Fig. 5d), which we believe promoted the growth and stability of MNs throughout the culture period; however, this trend was not observed for GDNF (Fig. 5e).
In the PNS, fully mature myelinating SCs express peripheral myelin protein 22 (PMP22) and myelin protein zero (P0) when compact myelin is formed. PMP22 and P0 are known to regulate myelin formation, thickness and integrity30,31. In the MN–SC coculture myelination model, abundant PMP22- and P0-expressing mature SCs along the axonal segments were clearly observed on day 21 when they were immunolabeled with NF-H, whereas the myelination process in cultures treated with GGF was blocked and showed decreased PMP22 and P0 expression (Fig. 5f–i). Thus, we demonstrated that supplementing AA in the MN–SC coculture medium allowed the maturation of SCs into myelinating cells to initiate wrapping and formation of the myelin sheath, whereas GGF treatment of the MN–SC coculture inhibited this process.
Finally, formation of nodes of Ranvier in the myelination model were visualized by observing axonal organization in the perinodal regions using contactin-associated protein (Caspr) immunocytochemistry27,32,33,34,35,36,37,38,39,40,41,42,43,44,45. On day 21 of the MN–SC coculture, Caspr-positive clusters were observed between the myelinated segments along the axons, representing formation of nodes of Ranvier (Fig. 5j). The presence of Caspr indicates that the nodes were mature in terms of their physiological morphologies, thus validating the in vitro myelination model in a 3D culture platform. The structure of the myelin sheath in our in vitro chip were further visualized using TEM cross-sectional images. In the MN monoculture, the presence of axons was observed throughout the culture (Supplementary Fig. 3). In the MN–SC coculture, unmyelinated axons with encapsulation of neurites by SCs were observed on day 7. On days 14 and 21, myelinating SCs were observed to be closely associated with axonal fibers (Supplementary Fig. 3). The presence of myelin sheaths was reduced in the MN–SC coculture group treated with GGF. Overall, our model showed both myelin formation and the development of nodes of Ranvier in ECM hydrogels and suggested that the cell culture setup in this study can serve as a tool to investigate the interaction between axons and SCs during myelination and demyelination.
Conclusion
An important aim of this study was to demonstrate an in vitro platform for MN–SC coculture to represent a model of the PNS and to assess the myelination process in a 3D environment. We proposed a robust method for modeling PNS myelination using a MN–SC coculture in a microengineered platform. This platform can be easily used to facilitate the coculture of different types of cells for bioengineering applications. Additionally, it can be used as a tool to assess axon–SC interactions during myelination and evaluate myelin formation in a more physiologically relevant environment than traditional 2D cell culture settings. In addition, we further showed the application of our model in disease modeling by demonstrating its capabilities to enable the long-term culture of MNs and SCs to expand scientific knowledge regarding the progressive stages of peripheral myelination, particularly in the study of motor neuron diseases.
References
Garbay, B., Heape, A. M., Sargueil, F. & Cassagne, C. Myelin synthesis in the peripheral nervous system. Prog. Neurobiol. 61, 267–304 (2000).
Salzer, J. L. Schwann cell myelination. Cold Spring Harb. Perspect. Biol. 7, 1–26 (2015).
Jessen, K. R. & Mirsky, R. The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 6, 671–682 (2005).
Salzer, J. L., Brophy, P. J. & Peles, E. Molecular domains of myelinated axons in the peripheral nervous system. Glia 56, 1532–1540 (2008).
Callizot, N., Combes, M., Steinschneider, R. & Poindron, P. A new long term in vitro model of myelination. Exp. Cell Res. 317, 2374–2383 (2011).
Stettner, M. et al. A reliable in vitro model for studying peripheral nerve myelination in mouse. J. Neurosci. Methods 214, 69–79 (2013).
Hyung, S. et al. Coculture of primary motor neurons and Schwann cells as a model for in vitro myelination. Sci. Rep. 5, 15122 (2015).
Shin, Y. et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 7, 1247–1259 (2012).
van Duinen, V., Trietsch, S. J., Joore, J., Vulto, P. & Hankemeier, T. Microfluidic 3D cell culture: from tools to tissue models. Curr. Opin. Biotechnol. 35, 118–126 (2015).
Eldridge, C. F., Bunge, M. B., Bunge, R. P. & Wood, P. M. Differentiation of axon-related Schwann cells in vitro. I. Ascorbic acid regulates basal lamina assembly and myelin formation. J. Cell Biol. 105, 1023–1034 (1987).
Laszkiewicz, I., Wiggins, R. C. & Konat, G. Ascorbic acid upregulates myelin gene expression in C6 glioma cells. Metab. Brain Dis. 7, 157–164 (1992).
Zanazzi, G. et al. Glial growth factor/neuregulin inhibits Schwann cell myelination and induces demyelination. J. Cell Biol. 152, 1289–1299 (2001).
Wiese, S. et al. Isolation and enrichment of embryonic mouse motoneurons from the lumar spinal cord of individual mouse embryos. Nat. Protoc. 5, 31–38 (2010).
Toy, D. & Namgung, U. Role of glial cells in axonal regeneration. Exp. Neurobiol. 22, 68–76 (2013).
Mitchel, J. A. & Hoffman-Kim, D. Cellular scale anisotropic topography guides Schwann cell motility. PLoS ONE 6, e24316 (2011).
Lopez-Fagundo, C., Bar-Kochba, E., Livi, L. L., Hoffman-Kim, D. & Frank, C. Three-dimensional traction forces of Schwann cells on compliant substrates. J. R. Soc. Interface 11, 20140247 (2014).
Edmondson, R., Broglie, J. J., Adcock, A. F. & Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay. Drug Dev. Technol. 12, 207–218 (2014).
Bunge, R. P. Tissue culture observations relevant to the study of axon–Schwann cell interactions during peripheral nerve development and repair. J. Exp. Biol. 132, 21–34 (1987).
Fawcett, J. W. & Keynes, R. J. Peripheral nerve regeneration. Annu. Rev. Neurosci. 13, 43–60 (1990).
Bunge, R. P. The role of the Schwann cell in trophic support and regeneration. J. Neurol. 242, S19–S21 (1994).
Yoshimura, K. & Takeda, S. Hedgehog signaling regulates myelination in the peripheral nervous system through primary cilia. Differentiation 83, S78–S85 (2012).
Feltri, M. L., Poitelon, Y. & Previtali, S. C. How Schwann cells sort axons: new concepts. Neuroscientist 22, 252–265 (2016).
Thompson, D. M. & Buettner, H. M. Neurite outgrowth is directed by Schwann cell alignment in the absence of other guidance cues. Ann. Biomed. Eng. 34, 669–676 (2006).
Heermann, S. & Schwab, M. H. Molecular control of Schwann cell migration along peripheral axons. Cell Adhes. Migr. 7, 18–22 (2013).
Liazoghli, D., Roth, A. D., Thostrup, P. & Colman, D. R. Substrate micropatterning as a new in vitro cell culture system to study myelination. ACS Chem. Neurosci. 3, 90–95 (2012).
Rumsey, J. W. et al. Node of Ranvier formation on motoneurons in vitro. Biomaterials 30, 3567–3572 (2009).
Poliak, S. & Peles, E. The local differentiation of myelinated axons at nodes of Ranvier. Nat. Rev. Neurosci. 4, 968–980 (2003).
Son, Y. J. & Thompson, W. J. Schwann cell processes guide regeneration of peripheral axons. Neuron 14, 125–132 (1995).
Scheib, J. & Hoke, Ahmet Advances in peripheral nerve regeneration. Nat. Rev. Neurol. 9, 668–676 (2013).
Martini, R. & Schachner, M. Molecular bases of myelin formation as revealed by investigations on mice deficient in glial cell surface molecules. Glia 19, 298–310 (1997).
Eichberg, J. Myelin P0: new knowledge and new roles. Neurochem. Res. 27, 1331–1340 (2002).
Fex Svenningsen, A., Shan, W. S., Colman, D. R. & Pedraza, L. Rapid method for culturing embryonic neuron-glial cell cocultures. J. Neurosci. Res. 72, 565–573 (2003).
Chan, J. R. et al. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron 43, 183–191 (2004).
Eshed, Y. et al. Gliomedin mediates Schwann cell-axon interaction and the molecular assembly of the nodes of Ranvier. Neuron 47, 215–229 (2005).
Liu, N., Varma, S., Shooter, E. M. & Tolwani, R. J. Enhancement of Schwann cell myelin formation by K252a in the Trembler-J mouse dorsal root ganglion explant culture. J. Neurosci. Res. 79, 310–317 (2005).
Honkanen, H. et al. Isolation, purification and expansion of myelination-competent, neonatal mouse Schwann cells. Eur. J. Neurosci. 26, 953–964 (2007).
Gingras, M., Beaulieu, M. M., Gagnon, V., Durham, H. D. & Berthod, F. In vitro study of axonal migration and myelination of motor neurons in a three-dimensional tissue-engineered model. Glia 56, 354–364 (2008).
Paivalainen, S. et al. Myelination in mouse dorsal root ganglion/Schwann cell cocultures. Mol. Cell Neurosci. 37, 568–578 (2008).
Rangaraju, S., Madorsky, I., Pileggi, J. G., Kamal, A. & Notterpek, L. Pharmacological induction of the heat shock response improves myelination in a neuropathic model. Neurobiol. Dis. 32, 105–115 (2008).
Feinberg, K. et al. A glial signal consisting of gliomedin and NrCAM clusters axonal Na+ channels during the formation of nodes of Ranvier. Neuron 65, 490–502 (2010).
Susuki, K. et al. Schwann cell spectrins modulate peripheral nerve myelination. Proc. Natl Acad. Sci. USA 108, 8009–8014 (2011).
Rumsey, J. W. et al. Myelination and node of Ranvier formation on sensory neurons in a defined in vitro system. In Vitro Cell Dev. Biol. Anim. 49, 608–618 (2013).
Huval, R. M. et al. Microengineered peripheral nerve-on-a-chip for preclinical physiological testing. Lab Chip 15, 2221–2232 (2015).
Sakai, K., Shimba, K., Kotani, K. & Jimbo, Y. A co-culture microtunnel technique demonstrating a significant contribution of unmyelinated Schwann cells to the acceleration of axonal conduction in Schwann cell-regulated peripheral nerve development. Integr. Biol. 9, 678–686 (2017).
Khoshakhlagh, P., Sivakumar, A., Pace, L. A., Sazer, D. W. & Moore, M. J. Methods for fabrication and evaluation of a 3D microengineered model of myelinated peripheral nerve. J. Neural. Eng. 15, 064001 (2018).
Acknowledgements
This work was supported by the National Research Foundation (NRF) of Korea grant funded by the Korean government (MSIT) (No. 2017 M3C7A1028866) and the Korea Evaluation Institute of Industrial Technology (KEIT) grant funded by the Korean government (MSIT) (No. 20009125).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, S.E., Ahn, J., Jeong, HE. et al. A three-dimensional in vitro model of the peripheral nervous system. NPG Asia Mater 13, 2 (2021). https://doi.org/10.1038/s41427-020-00273-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41427-020-00273-w