Abstract
Fungal infections cause ~1.5 million deaths each year worldwide, and the mortality rate of disseminated candidiasis currently exceeds that of breast cancer and malaria. The major reasons for the high mortality of candidiasis are the limited number of antifungal drugs and the emergence of drug-resistant species. Therefore, a better understanding of antifungal host defense mechanisms is crucial for the development of effective preventive and therapeutic strategies. Here, we report that DOCK2 (dedicator of cytokinesis 2) promotes indispensable antifungal innate immune signaling and proinflammatory gene expression in macrophages. DOCK2-deficient macrophages exhibit decreased RAC GTPase (Rac family small GTPase) activation and ROS (reactive oxygen species) production, which in turn attenuates the killing of intracellular fungi and the activation of downstream signaling pathways. Mechanistically, after fungal stimulation, activated SYK (spleen-associated tyrosine kinase) phosphorylates DOCK2 at tyrosine 985 and 1405, which promotes the recruitment and activation of RAC GTPases and then increases ROS production and downstream signaling activation. Importantly, nanoparticle-mediated delivery of in vitro transcribed (IVT) Rac1 mRNA promotes the activity of Rac1 and helps to eliminate fungal infection in vivo. Taken together, this study not only identifies a critical role of DOCK2 in antifungal immunity via regulation of RAC GTPase activity but also provides proof of concept for the treatment of invasive fungal infections by using IVT mRNA.
Similar content being viewed by others
Introduction
In recent years, fungal infections have become a deadly threat to human health. Infectious fungal diseases cause approximately one and a half million deaths every year worldwide, especially in individuals with conditions resulting in immunodeficiency, including immunosuppression, human immunodeficiency virus (HIV) infection, and primary immune deficiency (PID) [1, 2]. The global mortality rate of fungal infection currently exceeds that of breast cancer and malaria and is close to the rates associated with tuberculosis and HIV [1, 2]. Candida albicans is the most common fungal species isolated from infected patients and has become one of the leading causes of nosocomial blood infection [1, 3, 4]. Infections with multidrug-resistant C. auris have a mortality rate of ~60% and have spread worldwide in recent years [5]. During the coronavirus disease 2019 (COVID-19) pandemic, there were increasing cases of mucormycosis in patients with COVID-19, especially in India [6]. Overall, the limited number of antifungal drugs available in the clinic and emergent drug-resistant fungal species have led to great challenges in the effective control of invasive fungal infections.
C-type lectin receptors (CLRs) play a key role in the recognition of fungal infection by the innate immune system. CLRs include Dectin-1, Dectin-2/3, Mincle, and MRC1 (Mannose Receptor C-type 1), which recognize different components of the fungal cell wall, such as β-glucans, α-mannans, hyphal mannose, and glycolipids. These components are used to initiate the antifungal immune response [7]. In innate immune cells, such as macrophages and dendritic cells, a variety of CLRs recognize different components of the fungal cell wall and then activate spleen tyrosine kinase (SYK) and recruit the CARD9-BCL10-MALT1 (CBM) complex to activate the NF-κB and MAPK signaling pathways, which leads to the expression and secretion of proinflammatory cytokines, including tumor necrosis factor-α (TNF-α), IL-6, IL-1β and IL-23 [8,9,10]. One recent study found that JNK1 negatively regulates the activation of the antifungal signaling pathway by inducing the expression of CD23 [11]. Phagocytosis and reactive oxygen species (ROS) production are considered to play a critical role in the elimination of fungal infections [11, 12]. It has also been reported that Dectin-1 recruits RAC1 to produce ROS, which play an important role in the clearance of intracellular fungi [13]. In recent years, an increasing number of antifungal immune regulators have been identified, including PLC-γ2, PKC-δ, SHP-2, TAGAP, and EPHB2 [9, 10, 14,15,16]. In addition to the innate immune system, the adaptive immune system also plays an indispensable role in host defense against fungal infection. Th17 cells release IL-17, which activates IL-17 signaling to produce cytokines and chemokines and then recruits neutrophils for the elimination of fungal infections [9, 17,18,19,20]. In addition, protective antibodies have been found to play an indispensable role in antifungal immunity [21]. In recent years, some gene mutations related to fungal infection have been identified, such as mutations in CARD9, DECTIN-1, IL-17F, IL-17RA, STAT3, and TRAF3IP2, all of which lead to chronic candidiasis (chronic mucocutaneous candidiasis) [22,23,24,25]. Therefore, the innate immune system coordinates with the adaptive immune system in antifungal immunity.
DOCK2 is a member of the CDM protein family (CED-5 in Caenorhabditis elegans and DOCK180 in mammals) and is specifically expressed in hematopoietic cells [26]. It controls actin recombination and regulates lymphocyte activation, migration and morphological changes [26, 27]. Phosphatidic acid binds to the C-terminal region of DOCK2 and recruits DOCK2 to the cell membrane to mediate the chemotaxis of neutrophils [28]. In addition, as an atypical RAC activator, DOCK2 is involved in TLR-7- and TLR-9-mediated IFN-α expression in plasmacytic dendritic cells [29]. It has also been found that double allelic mutation of DOCK2 led to impairment of RAC1 activation in T cells of clinical patients and defects in chemokine-induced migration of T cells, B cells, and NK cells [30]. Therefore, it is generally believed that DOCK2 is the upstream molecule of RAC and functions as a guanine nucleotide exchange factor (GEF) for RAC. It has been reported that RAC1 is released from LyGDI under fungal stimulation, which promotes ROS production for the elimination of intracellular fungi [13]. However, the specific molecular mechanism of RAC1 activation in antifungal immunity has not been intensively studied.
In this study, we show that DOCK2-deficient macrophages exhibit decreased RAC1 activation and ROS production after fungal stimulation, which leads to attenuated inflammatory gene expression and fungicidal activity in macrophages. Mechanistically, activated SYK phosphorylates DOCK2 at tyrosine 985 and 1405, which promotes the recruitment and activation of RAC GTPases, thereby increasing ROS production for signaling activation and fungicidal activity in macrophages. Moreover, Dock2-deficient mice showed a more severe phenotype than wild-type (WT) mice in a fungemia model, which further confirms its critical role in antifungal immunity. Notably, the administration of nanoparticles carrying in vitro transcribed (IVT) Rac1 mRNA helped to specifically enhance antifungal immunity and accelerate the elimination of fungi in vivo, which provides proof of concept for the treatment of invasive fungal infections.
Results
DOCK2 is required for antifungal signaling activation in macrophages
In our previous study, we found that myosin 1f (MYO1F) plays an essential role in the activation of the antifungal innate immune signaling pathway by facilitating membrane-cytoplasmic trafficking of SYK and CARD9 [31]. As part of an ongoing effort to further understand the mechanism of the antifungal immune signaling pathway, we performed immunoprecipitation and mass spectrometry to identify MYO1F-interacting proteins. Interestingly, DOCK2 was identified by mass spectrometry as a MYO1F-interacting protein [31]. To explore the functional role of DOCK2 in antifungal signaling, we generated DOCK2 knockout (KO) THP-1 cells using CRISPR–Cas9 gene editing. Interestingly, we found that NF-κB and MAPK signaling pathway activation by heat-killed C. albicans (HKCA) or α-mannan was dramatically decreased in DOCK2-KO THP-1 cells compared to WT THP-1 cells (Fig. 1A). β-Glucans, which are Dectin-1 ligands on the cell wall, are exposed on HKCA cells [32]. Curdlan is a β-1,3-glucan isolated from Alcaligenes faecalis that is an agonist of Dectin-1 in immune cells [33]. Next, we examined whether DOCK2 is also involved in the Dectin-2/Dectin-3 signaling pathway by using stimulation with α-mannan, which is a Dectin-2/3-specific ligand. α-Mannan-induced NF-κB and MAPK signaling activation was also attenuated in DOCK2-KO THP-1 cells compared to WT control THP-1 cells (Fig. 1B). Next, we confirmed the essential role of DOCK2 in CLR signaling in mouse macrophages, and we found that the proinflammatory cytokine and chemokine expression induced by HKCA, α-mannan or curdlan was significantly reduced in Dock2-KO bone marrow-derived macrophages (BMDMs) compared to WT BMDMs (Fig. 1C–E). Furthermore, signaling activation induced by curdlan, HKCA, or α-mannan was dramatically reduced in Dock2-KO BMDMs compared to WT BMDMs (Fig. 1F–H). TNF-α and CXCL1 production was significantly reduced in Dock2-KO BMDMs compared to WT BMDMs after HKCA, curdlan, or α-mannan stimulation, as shown by ELISA (Fig. 1I). Together, these data indicate that DOCK2 plays a critical and conserved role in antifungal CLR signaling activation in humans and mice.
DOCK2 is required for antifungal signaling activation in macrophages. Wild-type THP-1 cells and DOCK2-KO THP-1 cells were stimulated with heat-killed C. albicans (MOI = 2) (A) or Mannan (100 μg/mL) (B) for the indicated times, followed by western blot analysis of the indicated proteins. BMDMs from WT mice or Dock2-KO mice were stimulated with heat-killed C. albicans (HKCA: heat-killed C. albicans, MOI = 2) (C), α-Mannan (100 μg/mL) (D) or Curdlan (100 μg/mL) (E) for the indicated times, followed by RT and real-time PCR analysis of the expression of the indicated genes. BMDMs from WT mice or Dock2-KO mice were stimulated with Curdlan (100 μg/mL) (F), Mannan (100 μg/mL) (G) or heat-killed C. albicans (MOI = 2) (H) for the indicated time, followed by western blot analysis of the indicated proteins. I BMDMs from WT mice or Dock2-KO mice were stimulated with heat-killed C. albicans (MOI = 2), Curdlan (100 μg/mL) or α-Mannan (100 μg/mL) for 24 h, followed by ELISA of the indicated cytokines. All error bars indicate the S.E.M of biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 based on a two-tailed unpaired t test (C–E, I). The data are representative of three independent experiments
DOCK2 plays a critical role in human antifungal immunity
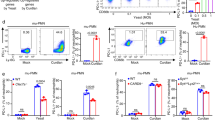
To further confirm the role of DOCK2 in human antifungal immunity, we knocked down the expression of DOCK2 in human peripheral blood mononuclear cells (PBMCs) isolated from the peripheral blood of healthy donors and examined whether DOCK2 played an important role in human primary cells. We found that knockdown of DOCK2 expression in human PBMCs significantly reduced the proinflammatory cytokine expression induced by curdlan, HKCA or mannan (Fig. 2A–C). These data further confirm our earlier data obtained in THP-1 cells and mouse BMDMs (Fig. 1).
DOCK2 plays a critical role in human antifungal immunity. PBMCs from healthy donors were transfected with control siRNA or DOCK2-siRNA, and 5 days after transfection were treated with Curdlan (100 μg/mL) (A), heat-killed C. albicans (HKCA, MOI = 2) (B) or Mannan (100 μg/mL) (C) for 3 h, followed by real-time PCR to analyze the expression of the indicated genes. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 based on one-way ANOVA in A–C. All error bars indicate the S.E.M of biological replicates. The data are representative of three independent experiments
DOCK2 mediates RAC GTPase activation and C. albicans clearance
Many studies have found that RAC GTPases are downstream molecules of DOCK2 that are crucial for activating ROS production via different pathways, such as the TCR signaling pathway [34]. However, whether DOCK2 can regulate RAC GTPase activation via CLR pathways has not been determined. Interestingly, we found that RAC1 activation was dramatically activated in WT THP-1 cells after HKCA or α-mannan stimulation but was almost completely abolished in DOCK2-KO THP-1 cells (Fig. 3A–B). These data indicate that DOCK2 controls RAC1 activation via CLR signaling pathways. ROS production is important for the clearance of intracellular pathogens, and we examined whether DOCK2 deficiency affects the clearance of intracellular fungi by human THP-1 cells. The results showed that the efficiency of live C. albicans clearance by DOCK2-KO THP-1 cells was significantly decreased compared to that by WT THP-1 cells and that overexpression of DOCK2 in THP-1 cells significantly increased C. albicans clearance compared to that achieved by control cells (Fig. 3C–E). DOCK2 functions as a GEF for RAC GTPases, and we explored whether this exchange activity of DOCK2 plays an important role in its antifungal activity. By in vitro fungal killing assays, we found that restoration of WT DOCK2 in DOCK2 KO THP-1 cells rescued antifungal activity, while restoration of a GEF activity-dead DOCK2 mutant (DOCK2-V1538A) failed to rescue antifungal activity in THP-1 cells. These data indicate that the guanine nucleotide exchange activity of DOCK2 is essential for antifungal immunity (Fig. 3F). Many studies have found that RAC GTPase activation is crucial for pathogen-induced ROS production and the activation of the bactericidal ability of macrophages [13], while it is still unclear whether RAC GTPase activation regulates antifungal signaling pathway activation and fungicidal activity. To further understand the role of RAC GTPase in the activation of the antifungal signaling pathway, we utilized the RAC GTPase inhibitor NSC23766 to determine whether RAC GTPase activation is necessary for antifungal signaling activation. The immunoblot results showed that curdlan-, α-mannan- or HKCA-induced signaling activation was dramatically decreased after pretreatment with the RAC GTPase inhibitor NSC23766 (Fig. 3G–I). Furthermore, curdlan-, α-mannan- or HKCA-induced proinflammatory cytokine and chemokine gene expression was significantly reduced after pretreatment with NSC23766 (Fig. 3J–K). Consistent with these findings, CLR-induced TNF-α and CXCL1 production was significantly reduced after pretreatment with NSC23766 (Fig. 3L). We examined cell survival after RAC inhibitor treatment and found that it was comparable between the NSC23766 treatment and control treatment groups; these data indicate that the cytokine production defects observed after NSC23766 treatment are not due to the cytotoxic effect of NSC23766 (Fig. 3M). A previous report indicated that DOCK2 also activates RAC2 [34]; thus, we further examined whether RAC2 is activated after fungal stimulation and whether its activity also relies on DOCK2. Interestingly, RAC2 was also activated after HKCA or mannan stimulation, and RAC2 activation was significantly reduced in DOCK2-KO THP-1 cells (Fig. S1A–B). These data indicate that fungus-induced RAC2 activation also relies on DOCK2 and that RAC2 may also contribute to fungus-induced ROS production and downstream signaling activation. Together, these data indicate that DOCK2 controls RAC GTPase activation and ROS production after CLR stimulation to eliminate fungal infection and inhibit downstream signaling activation.
DOCK2 is required for RAC1 activation and C. albicans clearance. WT THP-1 cells and DOCK2-KO THP-1 cells were stimulated with heat-killed C. albicans (MOI = 2) (A, left) or Mannan (100 μg/ml) (B, left) for the indicated times, followed by a RAC1 activity assay as described in the Methods section. Densitometric analysis of data from three independent experiments was performed for heat-killed C. albicans (MOI = 2) (A, right) and α-Mannan (100 μg/mL) (B, right) stimulation. C, D WT THP-1 cells and DOCK2-KO THP-1 cells were mixed with equal amounts of live C. albicans (MOI = 1) and cocultured for 24 h, followed by crystal violet staining of C. albicans (C). Scale bar = 5 μm. The fungal burden (D) was determined as described in the Methods section. E WT THP-1 cells overexpressing empty vector or Flag-DOCK2 were mixed with equal amounts of live C. albicans (MOI = 1) and cocultured for 24 h, and the fungal burden was determined as described in the Methods section. F DOCK2-KO THP-1 cells reconstituted with empty vector, WT DOCK2 or DOCK2 V1538A were mixed with equal amounts of live C. albicans (MOI = 1) and cocultured for 24 h, and the fungal burden was determined as described in the Methods section. BMDMs from WT mice were pretreated with NSC23766 (10 μM) for 1 h, followed by stimulation with Curdlan (100 μg/mL) (G), Mannan (100 μg/mL) (H) or heat-killed C. albicans (MOI = 2) (I) for the indicated times. Cell lysates were analyzed by western blotting for the indicated proteins. BMDMs from WT mice were pretreated with NSC23766 (10 μM) for 1 h and were then stimulated with Curdlan (100 μg/mL) (J) or Mannan (100 μg/mL) (K) for the indicated times, followed by real-time PCR to analyze the expression of the indicated genes. L BMDMs from WT mice were pretreated with NSC23766 (10 μM) for 1 h and were then stimulated with HKCA (MOI = 2), Curdlan (100 μg/mL) or α-Mannan (100 μg/mL) for 24 h, followed by ELISA to measure the production of the indicated proteins. M BMDMs from WT mice were pretreated with NSC23766 (10 μM) for 1 h prior to stimulation with heat-killed C. albicans (MOI = 2) for the indicated times, and cell survival was examined by an MTT assay. *P < 0.05; **P < 0.01; ***P < 0.001 based on a two-tailed unpaired t test (A, B, D, E, F, J–M). All error bars indicate the S.E.M of biological replicates. The data are representative of three independent experiments
DOCK2 controls ROS production via the CLR pathway
RAC GTPase activation is known to promote ROS production in response to pathogen stimulation, and we found that RAC1 and RAC2 GTPase activation was attenuated in DOCK2 KO macrophages compared to WT cells after fungal stimulation. We suspected that DOCK2 deficiency might lead to a reduction in ROS production in response to fungal stimulation. Consistent with our hypothesis, we found that Curdlan-, Mannan- or HKCA-induced ROS production was significantly reduced in DOCK2 KO THP-1 cells compared to WT THP-1 cells (Fig. 4A–C). These results suggest that DOCK2 plays a critical role in ROS production in macrophages after fungal infection. Curdlan-, mannan- or HKCA-induced ROS production was attenuated by RAC GTPase inhibitor treatment, which further confirmed that fungal-induced ROS production was dependent on RAC GTPase activation (Fig. 4D). Interestingly, RAC GTPase inhibitor treatment did not totally block ROS production after fungal stimulation, suggesting that in addition to DOCK2-RAC activation-induced ROS production, RAC-independent and DOCK2-dependent ROS production occurs (Fig. 4D). Interestingly, THP-1 cells treated with NAC and NSC23766 showed comparable antifungal effects, and these data suggest that RAC GTPase-induced ROS play an essential role in antifungal activity (Fig. S1C). Restoration of WT DOCK2 but not the GEF activity-dead DOCK2 mutant (DOCK2-V1538A), rescued ROS production after curdlan, mannan or HKCA treatment, and these data indicate that the guanine nucleotide exchange activity of DOCK2 plays a critical role in fungal-induced ROS production (Fig. 4E). It is generally believed that ROS are essential for the killing and scavenging of intracellular fungi during fungal infection [35], while the role of ROS in regulating antifungal signaling activation is unclear. Curdlan-, α-mannan- or HKCA-induced signaling activation and proinflammatory gene expression were dramatically impaired after pretreatment with the ROS inhibitor NAC compared to the control treatment (Fig. 4F–I). CXCL1 and TNFα production induced by HKCA, mannan or curdlan treatment was also significantly reduced after NAC treatment compared to the control treatment (Fig. 4J). To further prove the functional role of ROS in antifungal immune signaling, we examined whether another ROS inhibitor, Troxol, can also inhibit antifungal immunity. Similar to NAC, Troxol significantly inhibited curdlan-, mannan- or HKCA-induced signaling activation and cytokine production in BMDMs (Fig. S2A, B). Troxol also inhibited cytokine production in mouse serum after fungal stimulation (Fig. S2C). Together, these results indicate that DOCK2- and RAC1-dependent ROS production plays an indispensable role in the activation of the antifungal signaling pathway.
ROS play a critical role in antifungal signaling activation. WT THP-1 cells and DOCK2-KO THP-1 cells were stimulated with Curdlan (100 μg/mL) (A), Mannan (100 μg/mL) (B) or heat-killed C. albicans (MOI = 2) (C) for the indicated times, followed by measurement of ROS production. D WT THP-1 cells were pretreated with NSC23766 (10 μM) for 1 h and were then stimulated with Curdlan (100 μg/mL), α-Mannan (100 μg/mL) or heat-killed C. albicans (MOI = 2) for 30 min, followed by measurement of ROS production via a fluorescent ROS probe. E DOCK2-KO THP-1 cells reconstituted with vector, WT DOCK2 or DOCK2 V1538A were stimulated with Curdlan (100 μg/mL), α-Mannan (100 μg/mL) or heat-killed C. albicans (MOI = 2) for 30 min, followed by measurement of ROS production via a fluorescent ROS probe. F BMDMs from WT mice were pretreated with NAC (10 μM) for 1 h, followed by stimulation with Curdlan (100 μg/mL), Mannan (100 μg/mL) or heat-killed C. albicans (MOI = 2) for the indicated times. Cell lysates were analyzed by western blotting for the indicated proteins. BMDMs from WT mice were pretreated with NAC (10 μM) for 1 h and were then stimulated with Curdlan (100 μg/mL) (G), Mannan (100 μg/mL) (H) or heat-killed C. albicans (MOI = 2) (I) for the indicated times, followed by real-time PCR to analyze the expression of the indicated genes. J BMDMs from WT mice were pretreated with NAC (10 μM) for 1 h and were then stimulated with Curdlan (100 μg/mL), α-Mannan (100 μg/mL) or heat-killed C. albicans (MOI = 2) for 24 h, followed by ELISA of the indicated proteins. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 based on a two-tailed unpaired t test (D, E, G–J) or two-way ANOVA (A–C). All error bars indicate the S.E.M of biological replicates. The data are representative of three independent experiments
Phosphorylation of DOCK2 by SYK is required for RAC1 activation and ROS production
SYK is a key molecule in the antifungal immune signaling pathway. Under fungal stimulation, SYK is recruited to the cell membrane via the ITAM domain of Dectin-1 and then further recruits the CBM complex for activation of the NF-κB and MAPK signaling pathways, which ultimately induces the production and secretion of proinflammatory cytokines and chemokines [8,9,10]. In addition, it has been reported that activation of SYK promotes ROS production [36]. However, the mechanism by which activated SYK affects the production of ROS remains unclear. As SYK is a tyrosine kinase, we explored the possibility that DOCK2 is phosphorylated by SYK. To test our hypothesis, we first examined whether fungal stimulation induces phosphorylation of endogenous DOCK2 in THP-1 cells. Interestingly, DOCK2 was strongly phosphorylated in human THP-1 cells after stimulation with HKCA, and this phosphorylation was greatly attenuated after treatment with the SYK inhibitor piceatannol, suggesting that endogenous DOCK2 phosphorylation after fungal stimulation is dependent on SYK (Fig. 5A, B). Consistent with these findings, a kinase assay indicated that DOCK2 was phosphorylated by WT SYK but not a kinase-dead SYK mutant, which demonstrates that DOCK2 functions downstream of SYK in the Dectin-1 pathway (Fig. 5C). SYK also phosphorylated the GEF activity-dead mutant of DOCK2 (Fig. 5D). The interaction between DOCK2 and SYK was confirmed by coimmunoprecipitation and colocalization assays (Fig. 5E). To identify the domain(s) of DOCK2 or SYK that are responsible for their interaction, we generated DOCK2 and SYK truncation mutants and found that the interaction between DOCK2 and SYK is dependent on the SH3 domain of DOCK2 and the C-terminus of SYK (Fig. S3A, B). Furthermore, the phosphorylation of the DOCK2-ΔSH3 truncation mutant was obviously reduced (Fig. S3C).
Phosphorylation of DOCK2 by SYK is required for RAC activation and ROS production. A WT THP-1 cells were stimulated with heat-killed C. albicans (MOI = 2) for the indicated times, and cell lysates were immunoprecipitated with an anti-DOCK2 antibody (with IgG as the control), followed by western blot analysis of the indicated proteins. B WT THP-1 cells were pretreated with piceatannol (10 μM) for 1 h and were then stimulated with heat-killed C. albicans (MOI = 2) for the indicated times. Cell lysates were immunoprecipitated with an anti-DOCK2 antibody (IgG as the control), followed by western blot analysis of the indicated proteins. C HEK293T cells were transfected with the indicated plasmids, and cell lysates were immunoprecipitated with an anti-Flag antibody, followed by immunoblot analysis of the indicated proteins. D–F HEK293T cells were transfected with the indicated plasmids. Two days after transfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and the eluates were subjected to western blotting to analyze the indicated proteins. G HeLa cells were transfected with the indicated plasmids, followed by immunofluorescence staining with anti-Flag or anti-HA antibodies. Scale bar = 10 μm. H Alignment of the conserved motif in DOCK2 from different species. Red, tyrosine residues in DOCK2 phosphorylated by SYK identified by mass spectrometry. I, J DOCK2-KO THP-1 cells were reconstituted with wild-type DOCK2 or the DOCK2-Y985 + 1405 F mutant and stimulated with heat-killed C. albicans (MOI = 2) or α-Mannan (100 μg/mL) for the indicated times, followed by a RAC1 activity assay as described in the Methods section (I). Reconstituted cells were mixed with equal amounts of live C. albicans (MOI = 1) and cocultured for 24 h, and the fungal burden was then determined as described in the Methods section (J). K DOCK2-KO THP-1 cells were reconstituted with wild-type DOCK2 or Y985 + 1405 F mutant DOCK2 and were then stimulated with heat-killed C. albicans (MOI = 2), Mannan (100 μg/mL) or Curdlan (100 μg/mL) for 30 min, followed by measurement of ROS production via a fluorescent ROS probe. L DOCK2-KO THP-1 cells were reconstituted with wild-type DOCK2 or Y985 + 1405 F mutant DOCK2 and were then analyzed by western blotting. M DOCK2-KO THP-1 cells were reconstituted with wild-type DOCK2 or the DOCK2-Y985 + 1405 F mutant and stimulated with heat-killed C. albicans (MOI = 2) or Mannan (100 μg/mL) for the indicated times, followed by western blot analysis of the indicated proteins. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 based on one-way ANOVA (J–K). All error bars indicate the S.E.M of biological replicates. The data are representative of three independent experiments
By overexpressing SYK and DOCK2 in 293T cells and performing mass spectrometry, we identified 4 tyrosine residues in DOCK2, namely, Y122, Y224, Y985 and Y1405, which were phosphorylated by SYK (Fig. S4A). To understand the biological function of DOCK2 phosphorylation by SYK, we generated DOCK2 mutants in which each of the four tyrosine residues was mutated to phenylalanine (DOCK2-Y122F, DOCK2-Y224F, DOCK2-Y985F and DOCK2-Y1405F). Interestingly, the Y985F and Y1405F mutations in DOCK2 largely abolished its phosphorylation by SYK, indicating that SYK mainly phosphorylates DOCK2 at Y985 and Y1405 (Figs. 5F and S4B). However, the double Y985F + Y1405F mutation in DOCK2 did not affect its interaction with SYK (Figs. 5G and S4C–F). Sequence alignment showed that these two sites were quite conserved among different species (Fig. 5H). To further understand the functional role of DOCK2 phosphorylation by SYK in the antifungal signaling pathway, we reconstituted DOCK2 expression in DOCK2-KO THP-1 cells with WT DOCK2 or DOCK2-Y985 + 1405 F. WT DOCK2 rescued RAC1 activation after HKCA or α-mannan stimulation, while DOCK2-Y985 + 1405 F could not restore RAC1 activation (Fig. 5I). Reconstitution with WT DOCK2 rescued fungicidal activity and ROS production in DOCK2-KO THP-1 cells relative to that in empty vector control cells after live C. albicans or HKCA stimulation; in contrast, compared to reconstitution with WT DOCK2, reconstitution with the DOCK2-Y985 + 1405 F mutant only partially rescued fungicidal activity, and ROS production after reconstitution with the DOCK2-Y985 + 1405 F mutant was comparable to that in empty vector control cells (Fig. 5J–L). Reconstitution with WT DOCK2 but not the DOCK2-Y985 + 1405 F mutant significantly rescued HKCA- or α-mannan-induced signaling activation in DOCK2-KO THP-1 cells (Fig. 5M). Interestingly, erk activation in DOCK2-Y985 + 1405 F mutant-reconstituted cells was not totally abolished after mannan stimulation, suggesting that a SYK-DOCK2-phosphorylation-independent erk activation pathway may be induced after Dectin-2/3 stimulation. Together, these data indicate that phosphorylation of DOCK2 at Y985 and Y1405 by SYK is critical for its functional activity.
Phosphorylated DOCK2 is required for the recruitment of RAC GTPases to the cell membrane
To explore the mechanism of RAC1 activation by DOCK2 phosphorylation, we first investigated whether the DOCK2-Y985 +1405 F mutant affects the binding between DOCK2 and RAC1. The interaction between DOCK2 and RAC1 was confirmed by coimmunoprecipitation and was dependent on the DHR2 domain of DOCK2 (Fig. S4G–H). The interaction between DOCK2 and RAC1 was increased after HKCA stimulation in THP-1 cells (Fig. 6A). Interestingly, the interaction between RAC1 and the DOCK2-Y1405F mutant was substantially reduced, and DOCK2-Y985 also showed a reduced interaction with RAC1 (Fig. 6B). The interaction of the DOCK2-Y985 + 1405 F double mutant with RAC1 was almost totally abolished (Fig. 6C). These results indicate that the phosphorylation of DOCK2 at Y985 and Y1405 is critical for its interaction with RAC1.
Phosphorylated DOCK2 recruits RAC1 for membrane localization. A WT THP-1 cells were stimulated with heat-killed C. albicans (MOI = 2) for the indicated times, and cell lysates were immunoprecipitated with an anti-DOCK2 antibody (IgG as the control), followed by western blot analysis of the indicated proteins. B, C HEK293T cells were transfected with the indicated plasmids. Two days after transfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and the eluate was subjected to western blotting to analyze the indicated proteins. D, E Immunoblot analysis of cytosolic and membrane proteins from THP-1 WT cells and DOCK2-KO THP-1 cells stimulated with heat-killed C. albicans (MOI = 2) for the indicated times. DOCK2-KO THP-1 cells were reconstituted with wild-type DOCK2 or the DOCK2-Y985 + 1405 F mutant and were then stimulated with heat-killed C. albicans (MOI = 2, F) or Mannan (100 μg/mL, G) for the indicated times. Immunoblot analysis of the indicated cytosolic and cell membrane proteins is shown. H WT and DOCK2-KO THP-1 cells were stimulated with heat-killed C. albicans (MOI = 2) for the indicated times, followed by immunofluorescence analysis of the indicated proteins. Scale bar = 10 μm. I DOCK2-KO THP-1 cells were reconstituted with wild-type DOCK2 or the DOCK2-Y985 + 1405 F mutant and were then stimulated with heat-killed C. albicans (MOI = 2) for the indicated times, followed by immunofluorescence analysis of the indicated proteins. Scale bar = 10 μm. The data are representative of three independent experiments
During neutrophil chemotaxis, intracellular DOCK2 dynamics are sequentially regulated by distinct phospholipids to localize RAC1 activation [28]. Therefore, we explored whether DOCK2 localizes to the cell membrane after HKCA stimulation. Consistent with our hypothesis, HKCA or α-Mannan stimulation induced DOCK2 translocation to the cell membrane; interestingly, RAC1 was also recruited for membrane localization, while RAC1 membrane localization was almost completely abolished in DOCK2-KO THP-1 cells (Fig. 6D, E). Reconstitution of DOCK2 expression in DOCK2-KO THP-1 cells with WT DOCK2 but not the DOCK2-Y985 + 1405 F mutant restored the correct membrane localization of RAC1 after HKCA or α-Mannan stimulation (Fig. 6F, G), and this result was further confirmed by the finding that DOCK2 and RAC1 were colocalized after HKCA stimulation. There was strong colocalization between DOCK2 and RAC1 in WT cells but not in DOCK2-Y985 + 1405F-reconstituted THP-1 cells (Fig. 6H, I). These data indicate that the phosphorylation of DOCK2 at Y985 and Y1405 is required for RAC1 membrane localization after fungal stimulation.
Dock2-KO mice were susceptible to fungal sepsis
Next, we examined whether Dock2-KO mice exhibit any defects in the antifungal response to systemic fungal infection. Consistent with the in vitro findings, Dock2-KO mice showed a more severe phenotype than WT control mice, as shown by the weight loss and survival curves (Fig. 7A, B). The fungal burden was significantly increased in the kidneys of Dock2-KO mice compared to WT mice, and histological staining and periodic acid-Schiff (PAS) staining indicated more severe inflammation and a higher fungal burden in the kidneys of Dock2-KO mice than in those of control mice (Fig. 7C–E). Macrophages and neutrophils play a critical role in antifungal innate immunity and function as the first line of defense to eliminate fungal infections [19, 37]. After fungal infection, macrophages produce cytokines and chemokines, such as CXCL1 and GM-CSF, to promote the recruitment of neutrophils and macrophages to eliminate the infection [38, 39]. As we found that Dock2-KO macrophages had defects in the production of proinflammatory cytokines and chemokines, we explored whether Dock2-KO mice exhibited any defects in local macrophage or neutrophil recruitment after fungal infection. Consistent with our hypothesis, there was significantly reduced macrophage and neutrophil infiltration in the kidneys of Dock2-KO mice compared to those of WT control mice after systemic C. albicans infection (Fig. 7F–G). The serum TNF-α and CXCL1 levels were significantly reduced in Dock2-KO mice compared to control mice (Fig. 7H).
Dock2-KO mice were susceptible to fungal sepsis. Female WT control mice and Dock2-KO mice were injected intravenously with live C. albicans (4 × 105 cfu/100 μL 1 × PBS), and mouse weight loss (A), mouse survival (B), and the fungal burden in the kidneys (C) are shown. n = 12 (WT) and 12 (KO) in A–C. D, E Mice were treated as in A, and sacrificed 48 h after C. albicans infection. The kidneys were fixed, and the sections were stained with hematoxylin and eosin (H&E, left panel) or periodic acid-Schiff (PAS, right panel). n = 4. Scale bar = 506 μm. F, G WT and Dock2-KO mice were injected intravenously with live C. albicans (4 × 105 cfu/100 μL 1 × PBS) and sacrificed 24 h after fungal infection. Kidney cells were analyzed by flow cytometry. n = 9 (WT) and 8 (KO) in F; n = 13 (WT) and 10 (KO) in G. H Mice were treated as in A and sacrificed 24 h after C. albicans infection. Serum was analyzed by ELISA for the indicated proteins. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 based on a two-tailed unpaired t test (C, D, F–H), two-way ANOVA (A) and the log-rank (Mantel–Cox) test (B). All error bars indicate the S.E.M of biological replicates. The data are representative of three independent experiments
Nanocarrier-delivered Rac1 mRNA has a therapeutic effect on fungemia
As we found that fungal infection-induced RAC1 activation and ROS production play a critical role in the defense against fungal infection, we investigated whether ROS agonists can enhance fungal elimination. fMLP is known to induce the production of ROS [40]. Consistent with these findings, pretreatment with fMLP enhanced HKCA-induced inflammatory gene expression and signaling activation (Fig. S5A, B). Moreover, administration of fMLP facilitated the elimination of fungal infection in mice, as demonstrated by the reduction in weight loss, increase in survival and decrease in the fungal burden in the kidneys (Fig. S5C–E). Macrophage and neutrophil infiltration in mouse kidneys was significantly increased after fMLP treatment (Fig. S5F–G). It was found that fMLP administration can induce neutrophil recruitment [41]; therefore, the antifungal effect of fMLP may also be due to increased recruitment of neutrophils for fungal killing. Together, these results indicate that targeting ROS may be a feasible approach for the treatment of fungal infections.
Inspired by the mRNA vaccine against COVID-19 and siRNA drugs approved by the FDA [42, 43], we explored the feasibility of specifically enhancing antifungal immunity by boosting Rac1 activity through the introduction of nanocarrier-delivered Rac1 single-stranded mRNA in vitro and in vivo. We synthesized mRac1 mRNA in vitro and packaged it into lipid nanoparticles (LNPs) (Fig. 8A). mRNA-loaded LNPs were prepared by the ethanol dilution method to obtain mRac1@LNPs [44]. We chose the optimized weight ratio of total lipids to mRac1 (20:1) for mRac1@LNP preparation for further experiments due to the higher mRac1 encapsulation efficiency (Fig. S6A), suitable surface potential and uniform particle size (Fig. S6B–D). The transmission electron microscopy (TEM) images confirmed that the mRac1@LNPs were spherical nanoparticles consistent with LNPs but with a slightly larger diameter (~100 nm) (Fig. S6E, F), which indicated an increased particle size resulting from mRac1 encapsulation. To ensure the feasibility of mRac1 transfection via mRac1@LNPs in vitro and in vivo, we tested the stability of mRac1@LNPs after incubation with 10% fetal bovine serum (FBS). We found that the particle sizes and PDI of the mRac1@LNPs and LNPs remained nearly constant over 48 h at 37 °C (Fig. S7A, B). We speculated that the mRac1@LNPs and LNPs would not cause hemolysis after intravenous administration in vivo, based on the results of the simulated hemolysis assay in vitro (Fig. S7C). Considering the potential application of mRac1@LNPs, the stability of mRac1@LNPs and LNPs was guaranteed for 1 week (Fig. S7D). By using BMDMs, we found that transfection of mRac1-LNPs significantly increased Rac1 activation compared to transfection of the control GFP@LNPs (Fig. 8B). Compared to GFP@LNP transfection, mRac1@LNP transfection significantly increased ROS production after curdlan, mannan or HKCA stimulation (Fig. 8C). These data demonstrate that LNPs can effectively maintain the stability of Rac1 mRNA and deliver it into cells in vitro and that LNP-delivered Rac1 mRNA can efficiently promote Rac1 activation and ROS production in vitro. Interestingly, LNP-delivered Rac1 mRNA could not rescue Rac1 activity in Dock2-KO macrophages (Fig. S8A, B), nor could it rescue ROS and cytokine production in Dock2-KO macrophages after curdlan, mannan or HKCA stimulation (Fig. S8C, D). Together, these data indicate that DOCK2 is essential for the full activation of Rac1 and that overexpression of Rac1 cannot compensate for the defect in antifungal activity caused by DOCK2 deficiency. Next, we explored whether the delivery of mRac1@LNPs has any therapeutic effect on candidemia in vivo. Compared to injection of control GFP@LNPs, injection of mRac1@LNPs significantly attenuated the severity of the disease, as indicated by the weight changes, increase in survival and decrease in the fungal burden in the kidneys (Fig. 8D–F). Notably, compared to control GFP@LNP treatment, injection of mRac1@LNPs significantly increased the infiltration of macrophages and neutrophils (Fig. 8G, H), suggesting that mRac1@LNPs specifically boost antifungal immunity in vivo. It has been reported that mRNA-LNPs can be phagocytosed by monocytes, macrophages, dendritic cells and hepatocytes in vivo [45, 46]. We examined whether monocytes and macrophages can phagocytose mRNA-LNPs in peripheral blood and found that ~15 and 25% of the mRNA-LNPs were phagocytosed by monocytes and macrophages, respectively, in peripheral blood (Fig. 8I). These data indicate that monocytes and macrophages can phagocytose mRNA@LNPs in vivo. Together, these data suggest that nanoparticle-delivered mRac1 mRNA is a feasible agent for the treatment of fungal infections and provide a proof of concept therapeutic method for future infectious diseases.
mRac1@LNPs have a therapeutic effect on candidemia. A Schematic of mRac1 mRNA. B BMDMs from WT mice were transfected with control GFP-LNPs or mRac1@LNPs for 48 h and were then stimulated with HKCA (MOI = 2) or Mannan (100 μg/ml) for the indicated times, followed by a Rac1 activity assay as described in the Methods section. C BMDMs from WT mice were transfected with control GFP@LNPs or mRac1@LNPs for 48 h and were then stimulated with Curdlan (100 μg/ml), Mannan (100 μg/ml) or HKCA (MOI = 2) for 30 min, followed by measurement of ROS production via a fluorescent ROS probe. D, E WT female mice were injected intravenously with live C. albicans (4 × 105 cfu/100 μL 1 × PBS), and injected intravenously with control GFP@LNPs (0.25 μg/g) or mRac1@LNPs (0.25 μg/g) 12 h later. Mouse weight loss (D) and mouse survival (E) are shown. n = 12 in D, E. F Female WT mice were treated as in D, and the fungal burden in the kidneys was determined as described in the Methods section. n = 12 in F. G, H WT mice were treated as in D and sacrificed 24 h after fungal infection. Kidney cells were analyzed by flow cytometry. n = 8. I Female WT mice were injected intravenously with control GFP@LNPs (0.25 μg/g) and sacrificed 24 h after injection, and peripheral blood was collected and analyzed by flow cytometry. J Schematic presentation of the proposed model: after fungal stimulation, DOCK2 is phosphorylated at Y985 and Y1405 by SYK, which helps to recruit and activate RAC for ROS production and signaling activation. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 based on a two-tailed unpaired t test (C, F–H), two-way ANOVA (D) and the log-rank (Mantel–Cox) test (E). All error bars indicate the S.E.M of biological replicates. The data are representative of three independent experiments
Discussion
In the current study, we show that DOCK2-dependent RAC1 activity is indispensable for the activation of the CLR signaling pathway and the clearance of intracellular fungi in vitro and in vivo (Fig. 8J). Moreover, we show that liposomal carrier-mediated delivery of IVT Rac1 mRNA has a significant therapeutic effect on fungemia, which provides proof of concept for the treatment of lethal infections caused by fungi and other pathogens in humans.
ROS have long been considered toxic molecules that not only are important for antimicrobial immunity but also cause collateral damage to tissues, cells, and organelles. In addition to their direct antimicrobial effect, NOX2-dependent ROS have many other functions, including regulating phagosome pH and K+ levels [47, 48], activating signal transduction cascades, and altering the expression of specific genes [49]. Mitochondria-produced ROS have been shown to be involved in starvation-induced autophagy and autophagic cell death in cancer cells [50, 51]. In the antifungal CLR signaling pathway, ROS production plays a critical role in the clearance of intracellular fungi, while its role in signaling activation is still unknown. Here, we found that dectin-induced ROS are essential for the activation of dectin signaling pathways (Fig. 4E–I). The mechanism by which ROS regulate CLR signaling activation is still unknown and needs to be further investigated in the future. In DOCK2-KO THP-1 cells, ROS production was severely impaired (Fig. 4A–C), while ROS production was not totally blocked after RAC GTPase inhibitor treatment (Fig. 4D). These data suggest that fungus-induced ROS production is partially dependent on the DOCK2-RAC GTPase pathway and that there is a RAC GTPase-independent and DOCK2-dependent mechanism of ROS production. Interestingly, THP-1 cells showed comparable loss of fungicidal activity after RAC GTPase inhibitor or ROS inhibitor treatment (Fig. S1C), and these data suggest that DOCK2-RAC GTPase-induced ROS production plays a critical role in antifungal activity. This point requires further investigation in the future.
One recent study found that JNK1 inhibitors promote the activation of the antifungal pathway for the elimination of intracellular fungi, suggesting that small molecule inhibitors targeting JNK1 may have developmental potential as antifungal drugs [11]. Two other studies found that CBLB is a E3 ubiquitin ligase for Dectin-1/2 that participates in antifungal immune regulation through ubiquitination of Dectin-1/2. These studies suggest that CBLB may be a potential target for enhancing antifungal immunity [12, 52]. Our previous study found that administration of the Sirt2 deacetylase inhibitors AGK2, AK-1 and AK-7 protected mice against both systemic and central nervous system C. albicans infections [31]. In this study, we found that the ROS activator fMLP promoted the antifungal immune response and protected mice in a model of systemic fungal infection, suggesting that small molecules targeting ROS may be potential agents for the elimination of fungal infection.
In recent years, in vitro synthesized RNA has shown great potential as a new drug class in a broad range of medical areas, especially in mRNA vaccines against COVID-19, which have achieved great success. RNA-based drugs have considerable advantages over traditional drugs, such as monoclonal antibody drugs and small molecule drugs; for example, antibody drugs can only target membrane proteins, which account for a small proportion of the whole proteome, while mRNA drugs or siRNA drugs can theoretically target all proteins. Compared to small molecule drugs, mRNA drugs and siRNA drugs have improved specificity. Through the development of an in vivo RNA delivery system and the progress in RNA modifications to avoid stimulating the immune response, RNA-based drugs or vaccines have achieved great success in treating genetic diseases and preventing infectious diseases [43, 53, 54]. IVT RNA-based drugs have also been developed to treat infectious diseases, although almost all of these efforts were aimed at developing mRNA vaccines against pathogens, such as mRNA vaccines against SARS-CoV-2. In this study, we provide proof of concept for the treatment of fungal infections by boosting a specific antifungal pathway through the delivery of nanoparticle-delivered IVT mRNA in vivo. We employed intravenous injection of mRNA-LNPs into mice in the current study, and we found that ~41% of the mRNA@LNP particles in peripheral blood were taken up by monocytes and macrophages, as these cells are typical phagocytes [55]. However, we cannot exclude the possibility that other cell types, such as neutrophils, dendritic cells and hepatocytes, may also phagocytose mRNA@LNPs and play a role in antifungal immunity [56, 57]. Although this study aimed to identify an approach to treat invasive fungal infections, theoretically, the concept and method could be used for the treatment of infectious diseases caused by other pathogens, such as bacteria or viruses, a possibility that could be explored in the future.
Materials and methods
Human PBMC study
Whole blood samples were collected from each study participant. Human PBMCs were isolated with Ficoll-PaqueTM PREMIUM (17-5442-02, GE Healthcare) from freshly collected peripheral venous blood from healthy controls according to the manufacturer’s instructions. Briefly, Ficoll-Paque medium was added to the centrifuge tube, and the diluted blood sample was layered onto the Ficoll-Paque medium solution. The tube was centrifuged at 400 × g for 30–40 min at 18–20 °C with the brake turned off. The upper layer containing plasma and platelets was aspirated using a sterile pipette, leaving the layer of mononuclear cells undisturbed at the interface. The layer of mononuclear cells was washed twice with 1× PBS.
For the siRNA knockdown assays, human PBMCs were transfected with RfectSP siRNA Transfection Reagent (BioGenerator Biotechnology) according to the manufacturer’s instructions. Five days after transfection, human PBMCs were stimulated with HKCA (MOI = 2) or α-mannan (100 μg/mL) and harvested for gene expression analysis.
For stimulation of human PBMCs, cells were stimulated with curdlan (100 μg/mL), HKCA (MOI = 2), or α-mannan (100 μg/mL). Cells were then harvested for gene expression analysis. This study followed the guidelines established by the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (project number: S309). All study participants signed a written informed consent form.
Mice
Dock2 gene KO mice were a kind gift from Professor Chaohong Liu at Tongji Medical College of Huazhong University of Science & Technology and Professor Yoshinori Fukui at Kyushu University [26]. Mice were housed under SPF conditions, and the experimental protocols were approved by the Institutional Animal Care and Use Committee of Huazhong University of Science and Technology (project number: S909). This study complied with all relevant ethical regulations for animal research.
Cells
293T and HeLa cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. THP-1 cells were maintained in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin and 1% sodium pyruvate.
Preparation of mouse BMDMs
BMDMs were obtained by differentiating bone marrow progenitors from the tibiae and femora of 6–8-week-old male or female mice in Iscove’s modified Dulbecco’s medium containing 20 ng/mL M-CSF, 10% heat-inactivated FBS (Invitrogen), 1 mM sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen) for 5–7 days. Cells were then replated in 6-well or 12-well plates 1 day before experiments.
Reagents
The anti-p-SYK (clone C87C1), anti-p-IκBα (clone 14D4), anti-p-JNK (clone 81E11), anti-HA (clone C29F4) and anti-Flag (clone D6W5B) antibodies were purchased from Cell Signaling Technology (cat nos. 2710, 2859, 4668, 3724 and 14793, respectively). The anti-HA (clone HA-7) and anti-Flag (clone M2) antibodies used for immunoprecipitation were purchased from Sigma (cat nos. H9658 and F1804, respectively). The anti-MYC antibody was purchased from GNI (GNI4110-MC). The anti-DOCK2 (clone E-7), anti-p-ERK (clone 12D4), anti-p-tyrosine (clone PY20), anti-HSP90 (clone AC-16) and anti-Actin (clone C-2) antibodies were purchased from Santa Cruz Biotechnology (cat nos. sc-365242, sc-81492, sc-508, sc-130068 and sc-8432, respectively). The anti-SYK, anti-ERK1/2, anti-JNK1, anti-GAPDH, and anti-FcRγ antibodies were purchased from ABclonal (cat nos. A2123, A4782, A0288, A19056, and A12889, respectively). The anti-RAC1 and anti-RAC2 antibodies were purchased from Proteintech (cat nos. 24072-1-AP and 60077-1-Ig, respectively). The anti-CD45 (clone 30-F11), anti-Ly6G (clone 1A8), anti-F4/80 (clone BM8) and anti-CD11b (clone M1/70) antibodies used for flow cytometry were purchased from Biolegend (cat nos. 103108, 127608, 123115, and 101212, respectively). A Zombie Violet Fixable Viability Kit was purchased from Biolegend (cat no. 423114). Curdlan was purchased from InvivoGen (cat no. tlrl-cud). α-Mannan was purchased from Sigma (cat no. M3640). NSC23766 and NAC were purchased from Topscience (cat nos. T6342 and TP1088, respectively). fMLP was purchased from MCE (HY-P0224). Recombinant mouse M-CSF protein was purchased from Peprotech (cat no. 315-02). Mouse CXCL1 and TNF-α ELISA kits were purchased from Biolegend (447507 and 430904, respectively). A DNA purification kit was obtained from Axygen. Inorganic pyrophosphatase, rNTPs, DNase I, E. coli PolyA Polymerase, Vaccinia capping enzyme and a Monarch RNA Cleanup Kit were obtained from New England Biolabs. The reversed-phase column was obtained from Agilent. Dilinoleylmethyl-4-dimethylaminobutyrate (DLin-MC3-DMA), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and 1,2-dimyristoyl-rac-glycero-3-methoxypoly(ethylene glycol)-2000 (DMG-PEG2000) were provided by A.V.T. (Shanghai) Pharmaceutical Co., Ltd. Cholesterol was obtained from Aladdin (Shanghai, China).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and were then permeabilized with PBS containing 0.3% Triton X-100 for 10 min. Prior to incubation with the primary antibody, the samples were incubated with 10% goat serum at room temperature for 1 h to block nonspecific staining. After 12 h of incubation with the primary antibody at 4 °C, the samples were washed three times with ice-cold PBS and further stained with fluorophore (Alexa 488 and Alexa 594)-conjugated secondary antibodies. After antibody staining, the samples were counterstained with DAPI and immersed in mounting medium before being sealed on a slide with nail polish. Sealed slides were analyzed using an Olympus FV3000 microscope with the companion software.
Immunoblotting and immunoprecipitation
Cells were harvested and lysed on ice in lysis buffer containing 0.5% Triton X-100, 20 mM HEPES (pH 7.4), 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 10 mM NaF, 2 mM dithiothreitol, 1 mM sodium orthovanadate, 2 mM EGTA, 20 mM aprotinin, and 1 mM phenylmethylsulfonyl fluoride for 30 min and were then centrifuged at 12,000 rpm for 15 min to obtain clarified lysates. For immunoprecipitation, cell lysates were incubated with 1 μg of antibody at 4 °C overnight prior to incubation with protein A-Sepharose or protein G-Sepharose beads for 2 h. The beads were washed four times with lysis buffer, and the precipitates were eluted with 2× sample buffer and immunoblotted with antibodies.
Lentivirus-mediated gene knockout in THP-1 cells
The pLentiCRISPR-GFP vector was used for CRISPR/Cas9-mediated gene KO in the THP-1 cell line. Briefly, a lentiviral vector expressing a gRNA was transfected together with packaging plasmids into HEK293T (ATCC) packaging cells. Then, 48 and 72 h after transfection, virus-containing supernatants were harvested and filtered through a 0.2 μm filter. Target cells were infected twice and sorted by flow cytometry. For some experiments, single cells were sorted into 96-well plates by flow cytometry for isolation of single clones. Isolated single clones were verified by western blot analysis and DNA sequencing.
Generation of stable cell lines
For lentivirus packaging, 6.0 μg of the pCDH-MSCV-MCS-EF1-copGFP-T2A-Puro plasmid harboring DOCK2-Flag or DOCK2-Y985 + 1405F-Flag, 3.0 μg of the pMD2.G plasmid, and 3.0 μg of the psPAX2 plasmid were cotransfected into HEK293T cells in a 10 cm culture plate (8 × 106 cells per plate). Twelve hours post transfection, the medium was changed to fresh DMEM supplemented with 10% FBS, 5 mg/mL penicillin and 10 mg/mL streptomycin. Forty-eight hours post transfection, supernatants containing lentiviral particles were collected, 8 mL of each supernatant was used for two rounds of infection of DOCK2−/− THP-1 cells (3 × 106 cells per plate). Infected cells were selected with a final puromycin concentration of 6 µg/mL for 72 h and were then collected by centrifugation (1000 × g, 5 min) and cultured. The positive cell lines were identified by immunoblotting with an anti-Flag antibody.
RT and Real-time PCR
Total RNA was extracted from the spinal cord with TRIzol according to the manufacturer’s instructions. One microgram of total RNA for each sample was reverse transcribed using SuperScript® II Reverse Transcriptase from Thermo Fisher Scientific. The resulting complementary DNA was analyzed by real-time PCR using SYBR Green Real-Time PCR Master Mix. All gene expression results are expressed as arbitrary units relative to the expression of Actb or GAPDH.
Flow cytometry
One day after infection with live C. albicans, mice were sacrificed and perfused with 1× PBS. Kidneys were homogenized in ice-cold tissue grinders and filtered through a 100 μm cell strainer, and the cells were collected by centrifugation at 400 × g for 5 min at 4 °C. Cell surface staining was performed for 30 min at 4 °C. Zombie Violet Fixable Viability reagent (1:400; Biolegend) was added to exclude dead cells. Flow cytometry data analysis was performed by using CytExpert.
RAC1 activity evaluation
A total of 8 × 106 WT THP-1 cells or DOCK2-KO THP-1 cells were seeded in 10-cm plates. After overnight serum starvation, cells were infected for 0, 15, and 30 min with α-Mannan (100 μg/mL) or HKCA (MOI = 2). Cells were washed twice with ice-cold 1× PBS and lysed in lysis buffer containing 50 mM Tris (pH 7.5), 10 mM MgCl2, 0.5 M NaCl, and 2% IGEPAL, as described in the Rac1/Cdc42 Activation Assay Kit manual (Upstate). The supernatants were incubated with GST-PBD-agarose beads at 4 °C for 3 h, washed three times in wash buffer (25 mM Tris (pH 7.5), 30 mM MgCl2, 40 mM NaCl) and resuspended in loading buffer for immunoblot analysis.
ROS production assay
For the ROS production assay, ROS levels were measured with a DHE (dihydroethidium) fluorescent probe according to the manufacturer’s instructions. Briefly, 2 × 105 WT THP-1 cells or DOCK2-KO THP-1 cells were differentiated with PMA for 2 days and were then washed twice with 1 × PBS. DHE was added to fresh medium at a final concentration of 1 μM, and the cells were incubated at 37 °C for 30 min before stimulation with Curdlan (100 μg/mL), Mannan (100 μg/mL) or HKCA (MOI = 2). ROS production was measured by flow cytometry.
Mass spectrometry identification of DOCK2 phosphorylation sites
For identification of the sites in DOCK2 phosphorylated by SYK, 293T cells were transfected with Flag-hDOCK2 and empty vector or Flag-hDOCK2 plus HA-hSYK and were then subjected to immunoprecipitation with an anti-Flag antibody 2 days after transfection. After washing four times with IP buffer, proteins were eluted with IP buffer containing 0.1% SDS and analyzed by mass spectrometry. Samples were reduced and alkylated in dithiothreitol (DTT) and iodoacetamide prior to trypsin digestion overnight. Digested samples were injected onto an Agilent Zorbax 300SB-C18 0.075 mm × 150 mm column in an Eksigent NanoLC system coupled to a Thermo LTQ-ETD Orbitrap mass spectrometer. An Advion TriVersa NanoMate was used as the nano-electrospray ionization source, and MS data were searched against the RefSeq human protein database with Sorcerer Sequest. The searched dataset was processed through the TPP (Trans-Proteomics Pipeline) and filtered with Peptide Prophet.
In vitro fungal killing assay
For the in vitro fungal killing assay, differentiated WT THP-1 cells or DOCK2-KO THP-1 cells (5 × 105 cells/well) were incubated with live C. albicans (MOI = 1) for 24 h. After coculture, cells were washed with 1 × PBS three times and were then resuspended in fresh medium containing amphotericin B (Sigma–Aldrich, V900919) at a final concentration of 30 μg/mL and cultured for 3 h at 37 °C. Cells were washed again three times in 1× PBS and were then lysed by the addition of 0.02% Triton X-100 (Sigma–Aldrich, T8787), and 100 mL of the suspension (1:1000 dilution) was spread on YPD plates. After incubation at 37 °C for 24 h, killing was evaluated by counting the Candida colonies.
Membrane and cytoplasmic fractionation
WT THP-1 cells and DOCK2-KO THP-1 cells were rinsed with cold 1× PBS three times and washed one time in hypotonic buffer (10 mM KCl, 1.5 mM MgCl2, and 10 mM Tris-HCl (pH 7.5)) supplemented with a protease inhibitor, incubated on ice in hypotonic buffer for 15 min and then pipetted up and down 5–10 times. The lysates were centrifuged at 4 °C for 5 min at 2500 × g to remove nuclei and cellular debris. Supernatants were centrifuged at 100,000 × g for 60 min at 4 °C to separate the cytosolic extracts (S100) and pellets (P100). The pellets (P100) were resuspended in lysis buffer volumes equal to those of the supernatants (S100), stored with the addition of 5× loading buffer, and analyzed by western blotting.
Synthesis of single-stranded mRac1 mRNA
In vitro transcription (IVT) using VSW-3 RNA polymerase [58] was carried out to prepare mRac1 mRNA. DNA fragments containing the 5′UTR sequence, Flag coding sequence, mRac1 coding sequence and 3′UTR sequence under the control of the VSW-3 RNA polymerase promoter [58] were inserted into the HhaI and XbaI sites in the pUC18 plasmid. Then, a pair of amplification primers (F-primer: 5′-CGTTGTAAAACGACGGCCAGTGC-3′ and R-primer: 5′- ATAAAATTAAGTTGCATCAAGCT-3′) were used for PCR amplification of the DNA template for IVT. PCR products were cleaned with a DNA purification kit. The IVT reaction (200 μL) contained 40 mM Tris-HCl (pH 8.0), 16 mM MgCl2, 5 mM DTT, 2 mM spermidine, 35 ng/μL template DNA, 1.5 U/μL RNase inhibitor, 0.2 μM inorganic pyrophosphatase, each of the four NTPs (4 mM) and 0.15 μM VSW-3 RNA polymerase. After incubation at 25 °C for 12 h, 40 units of DNase I were added to the reaction mixture and incubated for an additional 30 min at 37 °C to remove the template DNA. RNA products were purified with a Monarch® RNA Cleanup kit in a final volume of 200 μl. Linear RNA was capped (cap0) using Vaccinia capping enzyme (NEB) according to the manufacturer’s instructions. Polyadenosine tails were added to the capped linear transcripts using E. coli PolyA Polymerase (NEB) according to the manufacturer’s instructions, and fully processed mRNA was purified with a Monarch® RNA Cleanup Kit. mRNA was further purified through a 25 × 150 mm reversed-phase column with a particle size of 10 μm and pore size of 4000 A° (Agilent; part number: PL1212-3103) on an Agilent 1260 Series HPLC system (Agilent). The exact sequences of the mRNAs and the corresponding translated proteins are provided in Table S1.
Production of lipid-based mRac1 nanoparticles (mRac1@LNPs)
mRNA-loaded LNPs were produced by the ethanol dilution method. All lipids, namely, DLin-MC3-DMA, cholesterol, DSPC, and DMG-PEG2000, were dissolved in ethanol at a molar ratio of 50:10:38.5:1.5. mRac-1 diluted in 10 mM sodium citrate buffer (pH 4.5) was rapidly mixed with the solution of the above lipids at a volume ratio of 3:1 (aqueous:organic) by pipetting and vortexing. After incubation for 20 min at room temperature, the resulting nanoparticles (mRac1@LNPs) were diluted with 1× PBS (pH 7.4) buffer for characterization and mRNA transfection in vitro. For mRac-1 transfection in vivo, the freshly produced nanoparticles were dialyzed against 1× PBS to completely remove the ethanol.
Characterization of mRac1@LNPs
mRNA encapsulation by LNPs was evaluated by a gel retardation assay. The mRac1@LNPs were formulated at various weight ratios of total lipids to mRNA and were then electrophoresed on a 2% agarose gel in 1× TAE buffer at 100 V for 10 min. The mRNA was stained with Gel Red (Biosharp, China) and visualized with a ChemiDoc XRS+ imaging system (Bio–Rad, USA).
The particle size and zeta potential of mRac1@LNPs were analyzed using a Zetasizer Nano ZS90 instrument (Malvern, UK) after dilution with 1 × PBS. To observe the morphology of mRac1@LNPs, the mRac1@LNPs solution was deposited onto carbon TEM grids and imaged by TEM (HT7700, Hitachi, Japan).
The stability of mRac1@LNPs in vitro and in vivo was evaluated by size and PDI measurement. The particle size and PDI of mRac1@LNPs after incubation with 10% FBS at 37 °C or storage at 4 °C in PBS were analyzed as previously described. To test the biosafety of mRac1@LNPs in vivo, a hemolysis assay was conducted by incubation with red blood cells isolated from mice. The relative hemolysis of mRac1@LNPs or blank LNPs was analyzed based on normal saline as the negative control (0%) and 1% Triton X-100 as the positive control (100%).
Histopathology
For histopathological analyses, kidneys were fixed with 10% neutral-buffered formalin, processed according to standard procedures, embedded in paraffin, and sectioned. Two-micrometer-thick sections were stained with hematoxylin and eosin (H&E) and PAS. Renal inflammation was scored based on the H&E and PAS staining percentage (percentage of the renal parenchyma and/or pelvis involved with tubulointerstitial nephritis and/or pyelonephritis) as not significant (score 0), less than 10% (score 1), 10–25% (score 2), 25–50% (score 3), or greater than 50% (score 4). The intralesional fungal burden was scored based on PAS staining as not significant (score 0); scant presence, in less than 10% of inflammatory foci (score 1); mild to moderate presence, in 10–25% of inflammatory foci (score 2); moderate to significant presence, in 25–50% of inflammatory foci (score 3); or significant presence, in more than 50% of inflammatory foci (score 4).
Systemic C. albicans infection model
Live cells of C. albicans strain SC 5314 (4 × 105 yeast cells in 0.1 mL of 1× PBS buffer) were injected intravenously into 6- to 8-week-old littermate mice of different genotypes. Infected mice were monitored daily for weight loss and survival. The fungal burden in the kidneys was measured 2 days after infection. After the kidneys were collected, tissue homogenates were serially diluted and plated on yeast extract–peptone–dextrose agar. Fungal colony-forming units were counted 24 h after plating. For the in vivo mRNA@LNP treatment experiment, female WT mice were intravenously injected with live C. albicans (4 × 105 cfu/100 μL 1 × PBS), and 12 h later, the mice were intravenously injected with control GFP@LNPs (0.25 μg/g) or mRac1@LNPs (0.25 μg/g). For the fMLP treatment experiments, WT mice were intravenously injected with live C. albicans (4 × 105 cfu/100 μL 1 × PBS), and 12 h later, the mice were injected intraperitoneally with DMSO (50 μL) or fMLP (0.5 μg/g/50 μL).
Human PBMCs study
Whole blood samples were collected from healthy participants. Human PBMCs were isolated with Ficoll-PaqueTM PREMIUM (17-5442-02, GE Healthcare) from freshly collected peripheral venous blood from healthy donors. Briefly, Ficoll-Paque medium was added to the centrifuge tube, and the diluted blood sample was layered onto the Ficoll-Paque medium solution. The tube was centrifuged at 400 × g for 30 to 40 min at 18 °C to 20 °C with the brake turned off. The upper layer containing plasma and platelets was aspirated using a sterile pipette, leaving the layer of mononuclear cells undisturbed at the interface. The layer of mononuclear cells was washed twice with 1× PBS. For the siRNA knockdown assays, human PBMCs were transfected with RfectSP siRNA Transfection Reagent (BioGenerator Biotechnology) according to the manufacturer’s instructions. Five days after transfection, human PBMCs were stimulated with Curdlan (100 μg/mL), HKCA (MOI = 2) or α-mannan (100 μg/mL) and harvested for gene expression analysis. This study followed the guidelines established by the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology. All study participants signed a written informed consent form.
Statistics
The significance of differences between two groups was determined by unpaired two-tailed t test, multiple-group comparisons were performed using one-way ANOVA, the clinical scores and weight curves were analyzed by two-way ANOVA with multiple comparisons, and the survival rate was analyzed by using the log-rank (Mantel–Cox) test. P < 0.05 was considered to indicate significance. The results are shown as the means, and the error bars indicate the standard error of the mean (S.E.M) of biological or technical replicates as indicated in the figure legend.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv113.
Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018;360:739–42.
Brandt ME, Warnock DW. Epidemiology, clinical manifestations, and therapy of infections caused by dematiaceous fungi. J Chemother. 2003;15:36–47.
Husain S, Alexander BD, Munoz P, Avery RK, Houston S, Pruett T, et al. Opportunistic mycelial fungal infections in organ transplant recipients: emerging importance of non-Aspergillus mycelial fungi. Clin Infect Dis. 2003;37:221–9.
Horton MV, Nett JE. Candida auris infection and biofilm formation: going beyond the surface. Curr Clin Microbiol Rep. 2020;7:51–56.
Singh AK, Singh R, Joshi SR, Misra A. Mucormycosis in COVID-19: a systematic review of cases reported worldwide and in India. Diabetes Metab Syndr. 2021;15:102146.
Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–78.
Hara H, Ishihara C, Takeuchi A, Imanishi T, Xue L, Morris SW, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–29.
Strasser D, Neumann K, Bergmann H, Marakalala MJ, Guler R, Rojowska A, et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-delta to elicit Card9 adaptor-mediated innate immunity. Immunity. 2012;36:32–42.
Xu S, Huo J, Lee KG, Kurosaki T, Lam KP. Phospholipase Cgamma2 is critical for Dectin-1-mediated Ca2+ flux and cytokine production in dendritic cells. J Biol Chem. 2009;284:7038–46.
Zhao X, Guo Y, Jiang C, Chang Q, Zhang S, Luo T, et al. JNK1 negatively controls antifungal innate immunity by suppressing CD23 expression. Nat Med. 2017;23:337–46.
Wirnsberger G, Zwolanek F, Asaoka T, Kozieradzki I, Tortola L, Wimmer RA, et al. Inhibition of CBLB protects from lethal Candida albicans sepsis. Nat Med. 2016;22:915–23.
Wu W, Hsu YM, Bi L, Songyang Z, Lin X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat Immunol. 2009;10:1208–14.
Chen J, He R, Sun W, Gao R, Peng Q, Zhu L, et al. TAGAP instructs Th17 differentiation by bridging Dectin activation to EPHB2 signaling in innate antifungal response. Nat Commun. 2020;11:1913.
Deng Z, Ma S, Zhou H, Zang A, Fang Y, Li T, et al. Tyrosine phosphatase SHP-2 mediates C-type lectin receptor-induced activation of the kinase Syk and anti-fungal TH17 responses. Nat Immunol. 2015;16:642–52.
Sun W, Wang H, Hu H, Ma X, Zhang H, Chen J, et al. Cutting Edge: EPHB2 Is a Coreceptor for Fungal Recognition and Phosphorylation of Syk in the Dectin-1 Signaling Pathway. J Immunol. 2021;206:1419–23.
Gringhuis SI, Kaptein TM, Wevers BA, Theelen B, van der Vlist M, Boekhout T, et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1beta via a noncanonical caspase-8 inflammasome. Nat Immunol. 2012;13:246–54.
Gross O, Poeck H, Bscheider M, Dostert C, Hannesschläger N, Endres S, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–6.
Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nat Immunol. 2012;13:817–22.
LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–8.
Doron I, Leonardi I, Li XV, Fiers WD, Semon A, Bialt-DeCelie M, et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell. 2021;184:1017–31. e1014
Boisson B, Wang C, Pedergnana V, Wu L, Cypowyj S, Rybojad M, et al. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 2013;39:676–86.
Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, et al. Human dectin-1 deficiency and mucocutaneous fungal infections. N. Engl J Med. 2009;361:1760–7.
Glocker EO, Hennigs A, Nabavi M, Schäffer AA, Woellner C, Salzer U, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl J Med. 2009;361:1727–35.
Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–68.
Fukui Y, Hashimoto O, Sanui T, Oono T, Koga H, Abe M, et al. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412:826–31.
Guo X, Chen SY. Dedicator of Cytokinesis 2 in Cell Signaling Regulation and Disease Development. J Cell Physiol. 2017;232:1931–40.
Nishikimi A, Fukuhara H, Su W, Hongu T, Takasuga S, Mihara H, et al. Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science. 2009;324:384–7.
Gotoh K, Tanaka Y, Nishikimi A, Nakamura R, Yamada H, Maeda N, et al. Selective control of type I IFN induction by the Rac activator DOCK2 during TLR-mediated plasmacytoid dendritic cell activation. J Exp Med. 2010;207:721–30.
Dobbs K, Domínguez Conde C, Zhang SY, Parolini S, Audry M, Chou J, et al. Inherited DOCK2 Deficiency in Patients with Early-Onset Invasive Infections. N. Engl J Med. 2015;372:2409–22.
Sun W, Ma X, Wang H, Du Y, Chen J, Hu H, et al. MYO1F regulates antifungal immunity by regulating acetylation of microtubules. Proc Natl Acad Sci USA. 2021;118:e2100230118.
Gow NA, Netea MG, Munro CA, Ferwerda G, Bates S, Mora-Montes HM, et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J Infect Dis. 2007;196:1565–71.
Kim HS, Park KH, Lee HK, Kim JS, Kim YG, Lee JH, et al. Curdlan activates dendritic cells through dectin-1 and toll-like receptor 4 signaling. Int Immunopharmacol. 2016;39:71–78.
Sanui T, Inayoshi A, Noda M, Iwata E, Oike M, Sasazuki T, et al. DOCK2 is essential for antigen-induced translocation of TCR and lipid rafts, but not PKC-theta and LFA-1, in T cells. Immunity. 2003;19:119–29.
Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–9.
Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106:2543–50.
Ward RA, Vyas JM. The first line of defense: effector pathways of anti-fungal innate immunity. Curr Opin Microbiol. 2020;58:160–5.
De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013;121:4930–7.
Wicks IP, Roberts AW. Targeting GM-CSF in inflammatory diseases. Nat Rev Rheumatol. 2016;12:37–48.
Schepers E, Glorieux G, Dhondt A, Leybaert L, Vanholder R. Role of symmetric dimethylarginine in vascular damage by increasing ROS via store-operated calcium influx in monocytes. Nephrol Dial Transpl. 2009;24:1429–35.
Nick JA, Avdi NJ, Young SK, Knall C, Gerwins P, Johnson GL, et al. Common and distinct intracellular signaling pathways in human neutrophils utilized by platelet activating factor and FMLP. J Clin Invest. 1997;99:975–86.
Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–9.
Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl J Med. 2020;383:2603–15.
Rizvi F, Everton E, Smith AR, Liu H, Osota E, Beattie M, et al. Murine liver repair via transient activation of regenerative pathways in hepatocytes using lipid nanoparticle-complexed nucleoside-modified mRNA. Nat Commun. 2021;12:613.
Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release. 2015;217:345–51.
Yanez Arteta M, Kjellman T, Bartesaghi S, Wallin S, Wu X, Kvist AJ, et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc Natl Acad Sci USA. 2018;115:E3351–E3360.
Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313.
Diebold BA, Smith SM, Li Y, Lambeth JD. NOX2 as a target for drug development: indications, possible complications, and progress. Antioxid Redox Signal. 2015;23:375–405.
Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15.
Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–52.
Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. 2008;15:171–82.
Xiao Y, Tang J, Guo H, Zhao Y, Tang R, Ouyang S, et al. Targeting CBLB as a potential therapeutic approach for disseminated candidiasis. Nat Med. 2016;22:906–14.
Second RNAi drug approved. Nat Biotechnol. 2020;38:385.
Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl J Med. 2020;383:1920–31.
Ramishetti S, Landesman-Milo D, Peer D. Advances in RNAi therapeutic delivery to leukocytes using lipid nanoparticles. J Drug Target. 2016;24:780–6.
Fenton OS, Kauffman KJ, Kaczmarek JC, McClellan RL, Jhunjhunwala S, Tibbitt MW, et al. Synthesis and Biological Evaluation of Ionizable Lipid Materials for the In Vivo Delivery of Messenger RNA to B Lymphocytes. Adv Mater. 2017;29. https://doi.org/10.1002/adma.201606944.
Lin Z, Xi L, Chen S, Tao J, Wang Y, Chen X, et al. Uptake and trafficking of different sized PLGA nanoparticles by dendritic cells in imiquimod-induced psoriasis-like mice model. Acta Pharm Sin B. 2021;11:1047–55.
Xia H, Jiang Y, Cheng R, Yu B, Lu X, Wu H, et al. (2020) In vitro transcription using psychrophilic phage VSW-3 RNA polymerase. bioRxiv, https://doi.org/10.1101/2020.09.14.297226.
Acknowledgements
We thank Shuyan Liang and Zhixin Qiu from Wuhan Biobank Co., Ltd., for their kind help with flow cytometric analysis. This investigation was supported by a grant from the National Key Research and Development Program of China (2020YFA0710700 to CHW); the National Natural Science Foundation of China (Grant No. 82101859 to WWS); the Original Exploration Program of National Natural Science Foundation of China (82150102, to CHW); the Key Research and Development Program of Sichuan Province (22ZDYF3738, to CHW); the Fundamental Research Funds for the Central Universities, HUST (2021GCRC031 to CHW); the National Natural Science Foundation of China (31870165 to BZ) and the Shenzhen Science and Technology Innovation Fund (JCYJ20210324115811032 to BZ).
Author information
Authors and Affiliations
Contributions
XJM performed the experiments with assistance from WWS, HPW, HJH, YYD, RG, RRH, QWP, ZHC, TP, and XF; JHW and CQX helped to obtain human PBMC samples. BBY and BZ helped to synthesize the mRac-1 single-stranded mRNA; XT and WL helped to produce and characterize the mRac-1-Flag@LNPs; XJM and CHW analyzed the data; and WWS and CHW wrote the manuscript. CHW supervised the project with BZ and WL.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Ma, X., Tan, X., Yu, B. et al. DOCK2 regulates antifungal immunity by regulating RAC GTPase activity. Cell Mol Immunol 19, 602–618 (2022). https://doi.org/10.1038/s41423-021-00835-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-021-00835-0
Keywords
This article is cited by
-
Multiple Immune Defects in Two Patients with Novel DOCK2 Mutations Result in Recurrent Multiple Infection Including Live Attenuated Virus Vaccine
Journal of Clinical Immunology (2023)
-
GSH-responsive camptothecin prodrug-based hybrid micellar nanoparticles enable antitumor chemo-immunotherapy by PD-L1 knockdown
Nano Research (2023)