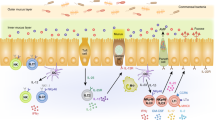
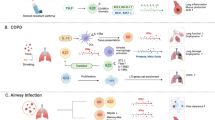
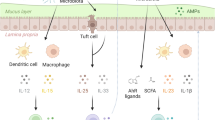
Abstract
The mucosal immune system is considered a local immune system, a term that implies regional restriction. Mucosal tissues are continually exposed to a wide range of antigens. The regulation of mucosal immune cells is tightly associated with the progression of mucosal diseases. Innate lymphoid cells (ILCs) are abundant in mucosal barriers and serve as first-line defenses against pathogens. The subtype changes and translocation of ILCs are accompanied by the pathologic processes of mucosal diseases. Here, we review the plasticity and circulation of ILCs in the mucosal immune system under physiological and pathological conditions. We also discuss the signaling pathways involved in dynamic ILC changes and the related targets in mucosal diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Spits, H. & Cupedo, T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30, 647–675 (2012).
Spits, H. et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145 (2013).
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018).
Warner, K. & Ohashi, P. S. ILC regulation of T cell responses in inflammatory diseases and cancer. Semin. Immunol. 41, 101284 (2019).
Fachi, J. L. et al. Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J. Exp. Med. 217, 1–18 (2020).
Seillet, C. et al. The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat. Immunol. 21, 168–177 (2020).
Wang, S. et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell 171, 201–16 e18 (2017).
Seehus, C. R. et al. Alternative activation generates IL-10 producing type 2 innate lymphoid cells. Nat. Commun. 8, 1900 (2017).
Cella, M. et al. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20, 980–991 (2019).
Rankin, L. C. et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol. 17, 179–186 (2016).
Miller, D. et al. Innate lymphoid cells in the maternal and fetal compartments. Front Immunol. 9, 2396 (2018).
Koues, O. I. et al. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell 165, 1134–1146 (2016).
Duffin, R. et al. Prostaglandin E(2) constrains systemic inflammation through an innate lymphoid cell-IL-22 axis. Science 351, 1333–1338 (2016).
Huang, Y. et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119 (2018).
Kobayashi, T. et al. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell 176, 982–97 e16 (2019).
Golebski, K. et al. IL-1beta, IL-23, and TGF-beta drive plasticity of human ILC2s towards IL-17-producing ILCs in nasal inflammation. Nat. Commun. 10, 2162 (2019).
Bjorklund, A. K. et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17, 451–460 (2016).
Bielecki, P. et al. Skin-resident innate lymphoid cells converge on a pathogenic effector state. Nature 592, 128–132 (2021).
Gury-BenAri, M. et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–46. e13 (2016).
Wang, S. et al. Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer. Cell Res. 30, 610–622 (2020).
Harly, C. et al. Development and differentiation of early innate lymphoid progenitors. J. Exp. Med. 215, 249–262 (2018).
Ohne, Y. et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol. 17, 646–655 (2016).
Lim, A. I. et al. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J. Exp. Med. 213, 569–583 (2016).
Silver, J. S. et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 17, 626–635 (2016).
Bal, S. M. et al. IL-1beta, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 17, 636–645 (2016).
Huang, Y. et al. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 16, 161–169 (2015).
Bernink, J. H. et al. c-Kit-positive ILC2s exhibit an ILC3-like signature that may contribute to IL-17-mediated pathologies. Nat. Immunol. 20, 992–1003 (2019).
van de Pavert, S. A. & Vivier, E. Differentiation and function of group 3 innate lymphoid cells, from embryo to adult. Int. Immunol. 28, 35–42 (2016).
Bernink, J. H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229 (2013).
Bernink, J. H. et al. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity 33, 736–751 (2010).
Mazzurana, L. et al. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3-ILC1/NK cell transdifferentiation. Eur. J. Immunol. 49, 1344–1355 (2019).
Parker, M. E. et al. c-Maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J. Exp. Med. 217, 1–22 (2020).
Koh, J. et al. IL23-producing human lung cancer cells promote tumor growth via conversion of innate lymphoid cell 1 (ILC1) into ILC3. Clin. Cancer Res. 25, 4026–4037 (2019).
Vacca, P. et al. Human natural killer cells and other innate lymphoid cells in cancer: friends or foes? Immunol. Lett. 201, 14–19 (2018).
Morita, H. et al. Induction of human regulatory innate lymphoid cells from group 2 innate lymphoid cells by retinoic acid. J. Allergy Clin. Immunol. 143, 2190–201 e9 (2019).
Howard, E. et al. IL-10 production by ILC2s requires Blimp-1 and cMaf, modulates cellular metabolism, and ameliorates airway hyperreactivity. J. Allergy Clin. Immunol. 147, 1281–1295 (2021).
Bando, J. K. et al. ILC2s are the predominant source of intestinal ILC-derived IL-10. J. Exp. Med. 217, 1–9 (2020).
Miyamoto, C. et al. Runx/Cbfbeta complexes protect group 2 innate lymphoid cells from exhausted-like hyporesponsiveness during allergic airway inflammation. Nat. Commun. 10, 447 (2019).
Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature 517, 293–301 (2015).
Almeida, F. F. & Belz, G. T. Innate lymphoid cells: models of plasticity for immune homeostasis and rapid responsiveness in protection. Mucosal Immunol. 9, 1103–1112 (2016).
Daussy, C. et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211, 563–577 (2014).
Bai, L. et al. Liver type 1 innate lymphoid cells develop locally via an interferon-gamma-dependent loop. Science. 371, 1–8 (2021).
Marcus, A. et al. Recognition of tumors by the innate immune system and natural killer cells. Adv. Immunol. 122, 91–128 (2014).
Malmberg, K. J. et al. Natural killer cell-mediated immunosurveillance of human cancer. Semin. Immunol. 31, 20–29 (2017).
Di Vito, C. et al. NK cells to cure cancer. Semin. Immunol. 41, 101272 (2019).
Gao, Y. et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017).
Cortez, V. S. et al. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-beta signaling. Nat. Immunol. 18, 995–1003 (2017).
Hawke, L. G. et al. TGF-beta and IL-15 synergize through MAPK pathways to drive the conversion of human NK cells to an innate lymphoid cell 1-like phenotype. J. Immunol. 204, 3171–3181 (2020).
Park, E. et al. Toxoplasma gondii infection drives conversion of NK cells into ILC1-like cells. Elife 8, 1–25 (2019).
Pikovskaya, O. et al. Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 196, 1449–1454 (2016).
Zhang, K. et al. Cutting edge: notch signaling promotes the plasticity of group-2 innate lymphoid cells. J. Immunol. 198, 1798–1803 (2017).
Flamar, A. L. et al. Interleukin-33 induces the enzyme tryptophan hydroxylase 1 to promote inflammatory group 2 innate lymphoid cell-mediated immunity. Immunity 52, 606–19 e6 (2020).
Chea, S. et al. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci. Signal. 9, ra45 (2016).
Rankin, L. C. et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14, 389–395 (2013).
Klose, C. S. et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature 494, 261–265 (2013).
Qi, X. et al. Brg1 restrains the pro-inflammatory properties of ILC3s and modulates intestinal immunity. Mucosal Immunol. 14, 38–52 (2021).
Teunissen, M. B. M. et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR(+) ILC3 in lesional skin and blood of psoriasis patients. J. Invest Dermatol. 134, 2351–2360 (2014).
Viant, C. et al. Transforming growth factor-beta and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 9, ra46 (2016).
Verrier, T. et al. Phenotypic and functional plasticity of murine intestinal NKp46+ group 3 innate lymphoid cells. J. Immunol. 196, 4731–4738 (2016).
Cortez, V. S. et al. Transforming growth factor-beta signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 44, 1127–1139 (2016).
Gasteiger, G. et al. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015).
Lim, A. I. et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell 168, 1086–100 e10 (2017).
Rao, A. et al. Cytokines regulate the antigen-presenting characteristics of human circulating and tissue-resident intestinal ILCs. Nat. Commun. 11, 2049 (2020).
Yang, J. et al. Selective programming of CCR10(+) innate lymphoid cells in skin-draining lymph nodes for cutaneous homeostatic regulation. Nat. Immunol. 17, 48–4 (2016).
Dutton, E. E. et al. Peripheral lymph nodes contain migratory and resident innate lymphoid cell populations. Sci. Immunol. 4, 1–14 (2019).
Kim, M. H. et al. Retinoic acid differentially regulates the migration of innate lymphoid cell subsets to the gut. Immunity 43, 107–119 (2015).
Vivier, E. et al. Functions of natural killer cells. Nat. Immunol. 9, 503–510 (2008).
Campbell, J. J. et al. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J. Immunol. 166, 6477–6482 (2001).
Frey, M. et al. Differential expression and function of L-selectin on CD56bright and CD56dim natural killer cell subsets. J. Immunol. 161, 400–408 (1998).
Lugthart, G. et al. Human lymphoid tissues harbor a distinct CD69+CXCR6+ NK cell population. J. Immunol. 197, 78–84 (2016).
Stegmann, K. A. et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci. Rep. 6, 26157 (2016).
Boning, M. A. L. et al. ADAP promotes degranulation and migration of NK cells primed during in vivo listeria monocytogenes infection in mice. Front. Immunol. 10, 3144 (2019).
Bajenoff, M. et al. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. J. Exp. Med. 203, 619–631 (2006).
Mayol, K. et al. Sequential desensitization of CXCR4 and S1P5 controls natural killer cell trafficking. Blood 118, 4863–4871 (2011).
Walzer, T. et al. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat. Immunol. 8, 1337–1344 (2007).
Morrison, B. E. et al. Chemokine-mediated recruitment of NK cells is a critical host defense mechanism in invasive aspergillosis. J. Clin. Invest. 112, 1862–1870 (2003).
Khan, I. A. et al. CCR5 is essential for NK cell trafficking and host survival following Toxoplasma gondii infection. PLoS Pathog. 2, e49 (2006).
Ajuebor, M. N. et al. CCR5 deficiency drives enhanced natural killer cell trafficking to and activation within the liver in murine T cell-mediated hepatitis. Am. J. Pathol. 170, 1975–1988 (2007).
Lavergne, E. et al. Fractalkine mediates natural killer-dependent antitumor responses in vivo. Cancer Res. 63, 7468–7474 (2003).
Wendel, M. et al. Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res. 68, 8437–8445 (2008).
Bernardini, G. et al. CCL3 and CXCL12 regulate trafficking of mouse bone marrow NK cell subsets. Blood 111, 3626–3634 (2008).
Martin-Fontecha, A. et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat. Immunol. 5, 1260–1265 (2004).
Wald, O. et al. IFN-gamma acts on T cells to induce NK cell mobilization and accumulation in target organs. J. Immunol. 176, 4716–4729 (2006).
Schneider, C. et al. Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and in situ perinatal priming. Immunity 50, 1425–38 e5 (2019).
Zeis, P. et al. In situ maturation and tissue adaptation of type 2 innate lymphoid cell progenitors. Immunity 53, 775–92 e9 (2020).
Ghaedi, M. et al. Single-cell analysis of RORalpha tracer mouse lung reveals ILC progenitors and effector ILC2 subsets. J. Exp. Med. 217, 1–19 (2020).
Ricardo-Gonzalez, R. R. et al. Tissue-specific pathways extrude activated ILC2s to disseminate type 2 immunity. J. Exp. Med. 217, 1–12 (2020).
Stier, M. T. et al. IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J. Exp. Med. 215, 263–281 (2018).
Karta, M. R. et al. beta2 integrins rather than beta1 integrins mediate Alternaria-induced group 2 innate lymphoid cell trafficking to the lung. J. Allergy Clin. Immunol. 141, 329–38 e12 (2018).
Xue, L. et al. Prostaglandin D2 activates group 2 innate lymphoid cells through chemoattractant receptor-homologous molecule expressed on TH2 cells. J. Allergy Clin. Immunol. 133, 1184–1194 (2014).
Salimi, M. et al. Group 2 innate lymphoid cells express functional NKp30 receptor inducing type 2 cytokine production. J. Immunol. 196, 45–54 (2016).
Oyesola, O. O. et al. The prostaglandin D2 receptor CRTH2 promotes IL-33-induced ILC2 accumulation in the lung. J. Immunol. 204, 1001–1011 (2020).
Ardain, A. et al. Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis. Nature 570, 528–532 (2019).
Satoh-Takayama, N. et al. The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity 41, 776–788 (2014).
Willinger, T. Oxysterols in intestinal immunity and inflammation. J. Intern. Med. 285, 367–380 (2019).
Chu, C. et al. Anti-microbial functions of group 3 innate lymphoid cells in gut-associated lymphoid tissues are regulated by G-protein-coupled receptor 183. Cell Rep. 23, 3750–3758 (2018).
Emgard, J. et al. Oxysterol sensing through the receptor GPR183 promotes the lymphoid-tissue-inducing function of innate lymphoid cells and colonic inflammation. Immunity 48, 120–32 e8 (2018).
Mackley, E. C. et al. CCR7-dependent trafficking of RORgamma(+) ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat. Commun. 6, 5862 (2015).
Pearson, C. et al. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. Elife 5, e10066 (2016).
Shikhagaie, M. M. et al. Neuropilin-1 is expressed on lymphoid tissue residing LTi-like group 3 innate lymphoid cells and associated with ectopic lymphoid aggregates. Cell Rep. 18, 1761–1773 (2017).
Li, J. et al. Aryl hydrocarbon receptor signaling involves in the human intestinal ILC3/ILC1 conversion in the inflamed terminal ileum of Crohn’s disease patients. Inflamm. Cell Signal. 3, 1–5 (2016).
Huot, N. et al. Natural killer cells migrate into and control simian immunodeficiency virus replication in lymph node follicles in African green monkeys. Nat. Med. 23, 1277–1286 (2017).
Germain, R. N. & Huang, Y. ILC2s—resident lymphocytes pre-adapted to a specific tissue or migratory effectors that adapt to where they move? Curr. Opin. Immunol. 56, 76–81 (2019).
Kloverpris, H. N. et al. Innate lymphoid cells are depleted irreversibly during acute HIV-1 infection in the absence of viral suppression. Immunity 44, 391–405 (2016).
Wang, X. et al. Innate lymphoid cell memory. Cell Mol. Immunol. 16, 423–429 (2019).
Cerwenka, A. & Lanier, L. L. Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol. 16, 112–123 (2016).
Liang, F. et al. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med. 9, 1–10 (2017).
Vaccari, M. et al. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat. Med. 22, 762–770 (2016).
Acknowledgements
This work was supported by the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDB29020000), the National Natural Science Foundation of China (81722023, 81922031), the Key Research Program of Frontier Sciences of the Chinese Academy of Sciences (ZDBS-LY-SM025), the Beijing Natural Science Foundation (7212067), and the Youth Innovation Promotion Association of CAS to S.W.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Shao, F., Yu, D., Xia, P. et al. Dynamic regulation of innate lymphoid cells in the mucosal immune system. Cell Mol Immunol 18, 1387–1394 (2021). https://doi.org/10.1038/s41423-021-00689-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-021-00689-6