Abstract
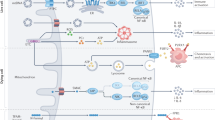
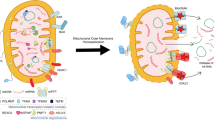
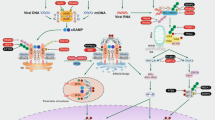
Mitochondria are highly mobile organelles due to fission, fusion, transport, and mitophagy, and these processes are known as mitochondrial dynamics. Mitochondrial dynamics play an important role in energy production, cell division, cell differentiation, and cell death. In the past decade, numerous studies have revealed the importance of mitochondrial metabolism in immunity, and mitochondrial dynamics are essential for immune responses mediated by various cell types. In this review, we mainly discuss the role of mitochondrial dynamics in activation, differentiation, cytokine production, and the activity of related pathways in immune cells, particularly T cells, B cells, and other cells involved in the innate immune response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Altieri, D. C. Mitochondrial dynamics and metastasis. Cell. Mol. Life Sci. 76, 827–835 (2019).
Mohanty, A., Tiwari-Pandey, R. & Pandey, N. R. Mitochondria: the indispensable players in innate immunity and guardians of the inflammatory response. J. Cell Commun. Signal. 13, 303–318 (2019).
El-Hattab, A. W., Suleiman, J., Almannai, M. & Scaglia, F. Mitochondrial dynamics: Biological roles, molecular machinery, and related diseases. Mol. Genet. Metab. 125, 315–321 (2018).
Mishra, P. Interfaces between mitochondrial dynamics and disease. Cell Calcium 60, 190–198 (2016).
Rambold, A. S. & Pearce, E. L. Mitochondrial dynamics at the interface of immune cell metabolism and function. Trends Immunol. 39, 6–18 (2018).
Baixauli, F. et al. The mitochondrial fission factor dynamin-related protein 1 modulates T-cell receptor signalling at the immune synapse. EMBO J. 30, 1238–1250 (2011).
Kang, Y. J. et al. Regulation of NKT cell-mediated immune responses to tumours and liver inflammation by mitochondrial PGAM5-Drp1 signalling. Nat. Commun. 6, 8371 (2015).
Zemirli, N., Morel, E. & Molino, D. Mitochondrial dynamics in basal and stressful conditions. Int. J. Mol. Sci. 19, 564 (2018).
Bulthuis, E. P., Adjobo-Hermans, M. J. W., Willems, P. & Koopman, W. J. H. Mitochondrial morphofunction in mammalian cells. Antioxid. Redox Signal 30, 2066–2109 (2019).
Gal, A. et al. MSTO 1 is a cytoplasmic pro‐mitochondrial fusion protein, whose mutation induces myopathy and ataxia in humans. EMBO Mol. Med. 9, 967–984 (2017).
Hoppins, S. The regulation of mitochondrial dynamics. Curr. Opin. Cell Biol. 29, 46–52 (2014).
Del Dotto, V., Fogazza, M., Carelli, V., Rugolo, M. & Zanna, C. Eight human OPA1 isoforms, long and short: What are they for? Biochim Biophys. Acta Bioenerg. 1859, 263–269 (2018).
Anand, R. et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204, 919–929 (2014).
Frezza, C. et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189 (2006).
Patten, D. A. et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33, 2676–2691 (2014).
Chan, D. C. Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. 15, 235–259 (2019).
Cho, B. et al. CDK5-dependent inhibitory phosphorylation of Drp1 during neuronal maturation. Exp. Mol. Med 46, e105 (2014).
Nakamura, N., Kimura, Y., Tokuda, M., Honda, S. & Hirose, S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 7, 1019–1022 (2006).
Gawlowski, T. et al. Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J. Biol. Chem. 287, 30024–30034 (2012).
Oliver, D. & Reddy, P. H. Dynamics of dynamin-related protein 1 in alzheimer’s disease and other neurodegenerative diseases. Cells. 8, 961 (2019).
Simula, L., Campanella, M. & Campello, S. Targeting Drp1 and mitochondrial fission for therapeutic immune modulation. Pharmacol. Res. 146, 8 (2019).
Burman, J. L. et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 216, 3231–3247 (2017).
Quintana, A. & Hoth, M. Mitochondrial dynamics and their impact on T cell function. Cell Calcium 52, 57–63 (2012).
Saxton, W. M. & Hollenbeck, P. J. The axonal transport of mitochondria. J. Cell Sci. 125, 2095–2104 (2012).
Pickles, S., Vigié, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 (2018).
Khaminets, A., Behl, C. & Dikic, I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 (2016).
Lazarou, M. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 (2015).
Palikaras, K., Lionaki, E. & Tavernarakis, N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022 (2018).
Twig, G. et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 (2008).
Weinberg, S. E., Sena, L. A. & Chandel, N. S. Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417 (2015).
Wang, R. & Green, D. R. Metabolic reprogramming and metabolic dependency in T cells. Immunol. Rev. 249, 14–26 (2012).
Buck, M. D., O’Sullivan, D. & Pearce, E. L. T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Klarquist, J., et al. Clonal expansion of vaccine-elicited T cells is independent of aerobic glycolysis. Sci. Immunol. 3, eaas9882 (2018).
Michalek, R. D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011).
Field, C. S. et al. Mitochondrial integrity regulated by lipid metabolism is a cell-intrinsic checkpoint for treg suppressive function. Cell Metab. 31, 422–437 e425 (2020).
Tan, H. et al. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 (2017).
Ma, E. H. et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8(+) T cells. Immunity 51, 856–870 e855 (2019).
Schwindling, C., Quintana, A., Krause, E. & Hoth, M. Mitochondria positioning controls local calcium influx in T cells. J. Immunol. 184, 184–190 (2010).
Quintana, A. et al. T cell activation requires mitochondrial translocation to the immunological synapse. Proc. Natl Acad. Sci. USA 104, 14418–14423 (2007).
Roth, D., Krammer, P. H. & Gulow, K. Dynamin related protein 1-dependent mitochondrial fission regulates oxidative signalling in T cells. FEBS Lett. 588, 1749–1754 (2014).
Krueger, A., Fas, S. C., Baumann, S. & Krammer, P. H. The role of CD95 in the regulation of peripheral T-cell apoptosis. Immunological Rev. 193, 58–69 (2003).
Serfling, E., Avots, A. & Neumann, M. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochimica et. Biophys. Acta 1263, 181–200 (1995).
Li-Weber, M., Laur, O., Dern, K. & Krammer, P. H. T cell activation-induced and HIV tat-enhanced CD95(APO-1/Fas) ligand transcription involves NF-kappaB. Eur. J. Immunol. 30, 661–670 (2000).
Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95 (2002).
Yu, T., Robotham, J. L. & Yoon, Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl Acad. Sci. USA 103, 2653–2658 (2006).
Cho, D.-H. et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Sci. (N. Y.) 324, 102–105 (2009).
Ron-Harel, N. et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 24, 104–117 (2016).
Klein Geltink, R. I. et al. Mitochondrial priming by CD28. Cell 171, 385–397 e311 (2017).
Simula, L. et al. Drp1 controls effective t cell immune-surveillance by regulating t cell migration, proliferation, and cMyc-dependent metabolic reprogramming. Cell Rep. 25, 3059–3073 e3010 (2018).
Zhao, G.-j et al. Up-regulation of mitofusin-2 protects CD4+ T cells from HMGB1-mediated immune dysfunction partly through Ca(2+)-NFAT signaling pathway. Cytokine 59, 79–85 (2012).
Caza, T. N. et al. HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann. Rheum. Dis. 73, 1888–1897 (2014).
Burté, F., Carelli, V., Chinnery, P. F. & Yu-Wai-Man, P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 11, 11–24 (2015).
Fan, K. Q. et al. Stress-induced metabolic disorder in peripheral CD4(+) T cells leads to anxiety-like behavior. Cell 179, 864–879 e819 (2019).
Hoffman, W., Lakkis, F. G. & Chalasani, G. B cells, antibodies, and more. Clin. J. Am. Soc. Nephrology: CJASN 11, 137–154 (2016).
Stein, M. et al. A defined metabolic state in pre B cells governs B-cell development and is counterbalanced by Swiprosin-2/EFhd1. Cell Death Differ. 24, 1239–1252 (2017).
Melchers, F. Checkpoints that control B cell development. J. Clin. Investig. 125, 2203–2210 (2015).
Sandoval, H., Kodali, S. & Wang, J. Regulation of B cell fate, survival, and function by mitochondria and autophagy. Mitochondrion 41, 58–65 (2018).
Boothby, M. & Rickert, R. C. Metabolic regulation of the immune humoral response. Immunity 46, 743–755 (2017).
Jin, G. et al. Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat. Immunol. 19, 29–40 (2017).
Wai, T. & Langer, T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27, 105–117 (2016).
Esteban-Martínez, L. et al. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 36, 1688–1706 (2017).
Zhang, Y. et al. Mitoguardin regulates mitochondrial fusion through mitopld and is required for neuronal homeostasis. Mol. Cell. 61, 111–124 (2016).
Gao, Z. et al. Mitochondrial dynamics controls anti-tumour innate immunity by regulating CHIP-IRF1 axis stability. Nat. Commun. 8, 1805 (2017).
Kano, S. et al. The contribution of transcription factor IRF1 to the interferon-gamma-interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat. Immunol. 9, 34–41 (2008).
Narayan, V., Pion, E., Landré, V., Müller, P. & Ball, K. L. Docking-dependent ubiquitination of the interferon regulatory factor-1 tumor suppressor protein by the ubiquitin ligase CHIP. J. Biol. Chem. 286, 607–619 (2011).
Yasukawa, K. et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci. Signal. 2, ra47 (2009).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015).
Yue, L. & Yao, H. Mitochondrial dysfunction in inflammatory responses and cellular senescence: pathogenesis and pharmacological targets for chronic lung diseases. Br. J. Pharm. 173, 2305–2318 (2016).
Kwon, D., Park, E. & Kang, S. J. Stimulator of IFN genes-mediated DNA-sensing pathway is suppressed by NLRP3 agonists and regulated by mitofusin 1 and TBC1D15, mitochondrial dynamics mediators. FASEB J. 31, 4866–4878 (2017).
Gkikas, I., Palikaras, K. & Tavernarakis, N. The role of mitophagy in innate immunity. Front. Immun. 9, 1283 (2018).
Bauernfeind, F. et al. Inflammasomes: current understanding and open questions. Cell Mol. Life Sci. 68, 765–783 (2011).
Kelley, N., Jeltema, D., Duan, Y. & He, Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 20, 3328 (2019).
Ichinohe, T., Yamazaki, T., Koshiba, T. & Yanagi, Y. Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proc. Natl Acad. Sci. USA 110, 17963–17968 (2013).
Subramanian, N., Natarajan, K., Clatworthy, M. R., Wang, Z. & Germain, R. N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 (2013).
Park, S. et al. Defective mitochondrial fission augments NLRP3 inflammasome activation. Sci. Rep. 5, 15489 (2015).
Wang, X. et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat. Immunol. 15, 1126–1133 (2014).
Kim, S. J., Ahn, D. G., Syed, G. H. & Siddiqui, A. The essential role of mitochondrial dynamics in antiviral immunity. Mitochondrion 41, 21–27 (2018).
Zhou, R., Yazdi, A. S., Menu, P. & Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011).
Zhong, Z. et al. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell 164, 896–910 (2016).
Li, S. et al. A novel mechanism of mesenchymal stromal cell-mediated protection against sepsis: restricting inflammasome activation in macrophages by increasing mitophagy and decreasing mitochondrial ROS. Oxid. Med. Cell Longev. 2018, 3537609 (2018).
Lupfer, C. et al. Receptor interacting protein kinase 2–mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 14, 480–488 (2013).
Ip, W. K. E., Hoshi, N., Shouval, D. S., Snapper, S. & Medzhitov, R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Sci. (N. Y.) 356, 513–519 (2017).
Misawa, T. et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454–460 (2013).
Wang, Y. et al. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell 171, 331–345 e322 (2017).
Krawczyk, C. M. et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115, 4742–4749 (2010).
Liu, Y.-J. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell. 106, 259–262 (2001).
Basit, F., Mathan, T., Sancho, D. & de Vries, I. J. M. Human dendritic cell subsets undergo distinct metabolic reprogramming for immune response. Front Immunol. 9, 2489 (2018).
Merad, M., Sathe, P., Helft, J., Miller, J. & Mortha, A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31, 563–604 (2013).
Du, X. et al. Hippo/Mst signalling couples metabolic state and immune function of CD8alpha(+) dendritic cells. Nature 558, 141–145 (2018).
Chang, C.-R. & Blackstone, C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 282, 21583–21587 (2007).
Cribbs, J. T. & Strack, S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8, 939–944 (2007).
Godfrey, D. I., Stankovic, S. & Baxter, A. G. Raising the NKT cell family. Nat. Immunol. 11, 197–206 (2010).
López-Armada, M. J., Riveiro-Naveira, R. R., Vaamonde-García, C. & Valcárcel-Ares, M. N. Mitochondrial dysfunction and the inflammatory response. Mitochondrion 13, 106–118 (2013).
West, A. P. Mitochondrial dysfunction as a trigger of innate immune responses and inflammation. Toxicology 391, 54–63 (2017).
Guilliams, M. et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578 (2014).
Duroux-Richard, I. et al. miR-125b controls monocyte adaptation to inflammation through mitochondrial metabolism and dynamics. Blood 128, 3125–3136 (2016).
Tondera, D. et al. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell Sci. 118, 3049–3059 (2005).
Abarca-Rojano, E. et al. Re-organization of mitochondria at the NK cell immune synapse. Immunol. Lett. 122, 18–25 (2009).
O’Sullivan, T. E., Johnson, L. R., Kang, H. H. & Sun, J. C. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity 43, 331–342 (2015).
Zheng, X. et al. Mitochondrial fragmentation limits NK cell-based tumor immunosurveillance. Nat. Immunol. 20, 1656–1667 (2019).
Angajala, A., et al. Diverse roles of mitochondria in immune responses: novel insights into immuno-metabolism. Front Immunol. 9, 1605 (2018).
Acknowledgements
This review was supported by the Excellent Young Scientist Foundation of NSFC (grant No. 31822017), the Zhejiang Provincial Natural Science Foundation of China under grant no. LR19C080001, the National Natural Science Foundation of China (grant nos. 81572651 and 81771675), and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
X.J. wrote the manuscript. J.J. and L.Y. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Xie, JH., Li, YY. & Jin, J. The essential functions of mitochondrial dynamics in immune cells. Cell Mol Immunol 17, 712–721 (2020). https://doi.org/10.1038/s41423-020-0480-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-020-0480-1
Keywords
This article is cited by
-
Targeting NEDD8 suppresses surgical stress-facilitated metastasis of colon cancer via restraining regulatory T cells
Cell Death & Disease (2024)
-
TREM-1 triggers necroptosis of macrophages through mTOR-dependent mitochondrial fission during acute lung injury
Journal of Translational Medicine (2023)
-
Integrative analysis of the prognostic value and immune microenvironment of mitophagy-related signature for multiple myeloma
BMC Cancer (2023)
-
OPA1 drives macrophage metabolism and functional commitment via p65 signaling
Cell Death & Differentiation (2023)
-
Potential role of mesenchymal stem cells in T cell aging
Journal of Molecular Medicine (2023)