Abstract
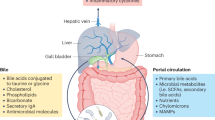
The gut microbiota is a complex and plastic consortium of microorganisms that are intricately connected with human physiology. The liver is a central immunological organ that is particularly enriched in innate immune cells and constantly exposed to circulating nutrients and endotoxins derived from the gut microbiota. The delicate interaction between the gut and liver prevents accidental immune activation against otherwise harmless antigens. Work on the interplay between the gut microbiota and liver has assisted in understanding the pathophysiology of various liver diseases. Of immense importance is the step from high-throughput sequencing (correlation) to mechanistic studies (causality) and therapeutic intervention. Here, we review the gut microbiota, liver immunology, and the interaction between the gut and liver. In addition, the impairment in the gut–liver axis found in various liver diseases is reviewed here, with an emphasis on alcohol-associated liver disease (ALD), nonalcoholic fatty liver disease (NAFLD), and autoimmune liver disease (AILD). On the basis of growing evidence from these preclinical studies, we propose that the gut–liver axis paves the way for targeted therapeutic modalities for liver diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
World Health Organization. The Top 10 Causes of Death. WHO, https://www.whoint/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (2018).
Younossi, Z. et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15, 11–20 (2018).
Seitz, H. K. et al. Alcoholic liver disease. Nat. Rev. Dis. Prim. 4, 16 (2018).
Wong, M. C. S. et al. The changing epidemiology of liver diseases in the Asia-Pacific region. Nat. Rev. Gastroenterol. Hepatol. 16, 57–73 (2019).
Mieli-Vergani, G. et al. Autoimmune hepatitis. Nat. Rev. Dis. Primers. 4, 18018 (2018).
Lleo, A. & Colapietro, F. Changes in the epidemiology of primary biliary cholangitis. Clin. Liver Dis. 22, 429–441 (2018).
Tripathi, A. et al. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411 (2018).
Chopyk, D. M. & Grakoui, A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology 159, 849–863 (2020).
Albillos, A. et al. The gut-liver axis in liver disease: pathophysiological basis for therapy. J. Hepatol. 72, 558–577 (2020).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
The Human Microbiome Project Consortium. et al. Structure, function and diversity of the healthy human microbiome. Nature. 486, 207–214 (2012).
Sender, R. et al. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Cheng, J. et al. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 10, 1002–1014 (2016).
Mariat, D. et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9, 123 (2009).
Ni, J. et al. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584 (2017).
Siljander, H. et al. Microbiome and type 1 diabetes. EBioMedicine 46, 512–521 (2019).
Gomes, A. C. et al. The human gut microbiota: metabolism and perspective in obesity. Gut Microbes 9, 308–325 (2018).
Tang, W. H. W. et al. Intestinal microbiota in cardiovascular health and disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 73, 2089–2105 (2019).
Fattorusso, A. et al. Autism spectrum disorders and the gut microbiota. Nutrients. 11, 521 (2019).
Gilbert, J. A. & Lynch, S. V. Community ecology as a framework for human microbiome research. Nat. Med. 25, 884–889 (2019).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
D’Haens, G. R. & Jobin, C. Fecal microbial transplantation for diseases beyond recurrent clostridium difficile infection. Gastroenterology 157, 624–636 (2019).
Nash, A. K. et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153 (2017).
Richard, M. L. & Sokol, H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 16, 331–345 (2019).
Standaert-Vitse, A. et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology 130, 1764–1775 (2006).
Coker, O. O. et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 68, 654–662 (2019).
Botschuijver, S. et al. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology 153, 1026–1039 (2017).
Lang, S. et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology 71, 522–538 (2020).
Lemoinne, S. et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 69, 92–102 (2020).
Shkoporov, A. N. & Hill, C. Bacteriophages of the human gut: The “Known Unknown” of the microbiome. Cell Host Microbe 25, 195–209 (2019).
Sausset, R. et al. New insights into intestinal phages. Mucosal Immunol. 13, 205–215 (2020).
Hoyles, L. et al. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 165, 803–812 (2014).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Shkoporov, A. N. et al. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 9, 4781 (2018).
Zuo, T. et al. Gut mucosal virome alterations in ulcerative colitis. Gut 68, 1169–1179 (2019).
Gogokhia, L. et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 25, 285–99.e8 (2019).
Nakatsu, G. et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155, 529–41.e5 (2018).
Kramná, L. et al. Gut virome sequencing in children with early islet autoimmunity. Diabetes Care 38, 930–933 (2015).
Międzybrodzki, R. et al. In vivo studies on the influence of bacteriophage preparations on the autoimmune inflammatory process. Biomed. Res. Int. 2017, 3612015 (2017).
Zuo, T. et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 67, 634–643 (2018).
Dong, X. et al. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 6, eaba1590 (2020).
Zheng, D. W. et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 3, 717–728 (2019).
Honda, K. & Littman, D. R. The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84 (2016).
Trompette, A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166 (2014).
Belkaid, Y. & Harrison, O. J. Homeostatic immunity and the microbiota. Immunity 46, 562–576 (2017).
Bunker, J. J. et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43, 541–553 (2015).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Ansaldo, E. et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019).
Ducarmon, Q. R. et al. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev. 83 (2019).
Brown, E. M. et al. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev. Immunol. 37, 599–624 (2019).
Shu, S. A. et al. Microbiota and food allergy. Clin. Rev. Allergy Immunol. 57, 83–97 (2019).
Roy, S. & Trinchieri, G. Microbiota: a key orchestrator of cancer therapy. Nat. Rev. Cancer 17, 271–285 (2017).
Racanelli, V. & Rehermann, B. The liver as an immunological organ. Hepatology 43, S54–S62 (2006).
Kolios, G. et al. Role of Kupffer cells in the pathogenesis of liver disease. World J. Gastroenterol. 12, 7413–7420 (2006).
Krenkel, O. & Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17, 306–321 (2017).
Wu, X. et al. Oral ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology 62, 253–264 (2015).
Rivera, C. A. et al. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J. Hepatol. 47, 571–579 (2007).
Vespasiani-Gentilucci, U. et al. Hepatic toll-like receptor 4 expression is associated with portal inflammation and fibrosis in patients with NAFLD. Liver Int. 35, 569–581 (2015).
Kawaratani, H. et al. Innate immune reactivity of the liver in rats fed a choline-deficient L-amino-acid-defined diet. World J. Gastroenterol. 14, 6655–6661 (2008).
Leroux, A. et al. Toxic lipids stored by Kupffer cells correlates with their pro-inflammatory phenotype at an early stage of steatohepatitis. J. Hepatol. 57, 141–149 (2012).
Kudo, H. et al. Lipopolysaccharide triggered TNF-alpha-induced hepatocyte apoptosis in a murine non-alcoholic steatohepatitis model. J. Hepatol. 51, 168–175 (2009).
Soderborg, T. K. et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat. Commun. 9, 4462 (2018).
Dapito, D. H. et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21, 504–516 (2012).
Mazagova, M. et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. Faseb j. 29, 1043–1055 (2015).
Krishnan, S. et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 23, 1099–1111 (2018).
Ma, L. et al. Indole alleviates diet-induced hepatic steatosis and inflammation in a manner involving myeloid cell 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3. Hepatology 72, 1191–1203 (2020).
Gong, S. et al. Intestinal microbiota mediates the susceptibility to polymicrobial sepsis-induced liver injury by granisetron generation in mice. Hepatology 69, 1751–1767 (2019).
McDonald, B. et al. Programing of an intravascular immune firewall by the gut microbiota protects against pathogen dissemination during infection. Cell Host Microbe 28, 660–668 (2020).
Hao, H. et al. Farnesoid X receptor regulation of the NLRP3 inflammasome underlies cholestasis-associated sepsis. Cell Metab. 25, 856–67.e5 (2017).
Isaacs-Ten, A. et al. Intestinal microbiome-macrophage crosstalk contributes to cholestatic liver disease by promoting intestinal permeability. Hepatology (2020).
Marrero, I. et al. Complex network of NKT cell subsets controls immune homeostasis in liver and gut. Front. Immunol. 9, 2082 (2018).
Bandyopadhyay, K. et al. NKT cell subsets as key participants in liver physiology and pathology. Cell Mol. Immunol. 13, 337–346 (2016).
Chen, J. et al. Natural killer T cells play a necessary role in modulating of immune-mediated liver injury by gut microbiota. Sci. Rep. 4, 7259 (2014).
Wei, Y. et al. Enterogenous bacterial glycolipids are required for the generation of natural killer T cells mediated liver injury. Sci. Rep. 6, 36365 (2016).
Llopis, M. et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65, 830–839 (2016).
Ma, C. et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 360, eaan5931 (2018).
Bonneville, M. et al. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10, 467–478 (2010).
Martin, B. et al. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity 31, 321–330 (2009).
Paget, C. et al. CD3bright signals on γδ T cells identify IL-17A-producing Vγ6Vδ1+ T cells. Immunol. Cell Biol. 93, 198–212 (2015).
Li, F. et al. The microbiota maintain homeostasis of liver-resident γδT-17 cells in a lipid antigen/CD1d-dependent manner. Nat. Commun. 7, 13839 (2017).
Tedesco, D. et al. Alterations in intestinal microbiota lead to production of interleukin 17 by intrahepatic γδ T-cell receptor-positive cells and pathogenesis of cholestatic liver disease. Gastroenterology 154, 2178–2193 (2018).
Provine, N. M. & Klenerman, P. MAIT cells in health and disease. Annu. Rev. Immunol. 38, 203–228 (2020).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 491, 717–723 (2012).
Legoux, F. et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366, 494–499 (2019).
Constantinides, M. G. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 366, eaax6624 (2019).
Lin, Q. et al. The dialogue between unconventional T cells and the microbiota. Mucosal Immunol. 13, 867–876 (2020).
Rha, M. S. et al. Human liver CD8(+) MAIT cells exert TCR/MR1-independent innate-like cytotoxicity in response to IL-15. J. Hepatol. 73, 640–650 (2020).
Niehaus, C. E. et al. MAIT cells are enriched and highly functional in ascites of patients with decompensated liver cirrhosis. Hepatology 72, 1378–1393 (2020).
Li, Y. et al. Mucosal-associated invariant T cells improve nonalcoholic fatty liver disease through regulating macrophage polarization. Front Immunol. 9, 1994 (2018).
Jiang, X. et al. The immunobiology of mucosal-associated invariant T cell (MAIT) function in primary biliary cholangitis: regulation by cholic acid-induced Interleukin-7. J. Autoimmun. 90, 64–75 (2018).
Böttcher, K. et al. MAIT cells are chronically activated in patients with autoimmune liver disease and promote profibrogenic hepatic stellate cell activation. Hepatology 68, 172–186 (2018).
Wahlström, A. et al. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016).
Jia, W. et al. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018).
Li, Y. et al. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun. Rev. 16, 885–896 (2017).
Tian, Y. et al. The microbiome modulating activity of bile acids. Gut Microbes 11, 979–996 (2020).
Sinha, S. R. et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 27, 659–70.e5 (2020).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature 577, 410–415 (2020).
Campbell, C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020).
Hang, S. et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 576, 143–148 (2019).
Glaser, F. et al. Liver infiltrating T cells regulate bile acid metabolism in experimental cholangitis. J. Hepatol. 71, 783–792 (2019).
Zarrinpar, A. & Loomba, R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Aliment Pharmacol. Ther. 36, 909–921 (2012).
Mouries, J. et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 71, 1216–1228 (2019).
Copple, B. L. & Li, T. Pharmacology of bile acid receptors: evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol. Res. 104, 9–21 (2016).
Nevens, F. et al. A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N. Engl. J. Med. 375, 631–643 (2016).
Gadaleta, R. M. et al. Fibroblast Growth Factor 19 modulates intestinal microbiota and inflammation in presence of Farnesoid X Receptor. EBioMedicine 54, 102719 (2020).
Liu, Y. et al. Probiotic Lactobacillus rhamnosus GG prevents liver fibrosis through inhibiting hepatic bile acid synthesis and enhancing bile acid excretion in mice. Hepatology 71, 2050–2066 (2020).
Fiorucci, S. & Distrutti, E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol. Med. 21, 702–714 (2015).
Foley, M. H. et al. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 15, e1007581 (2019).
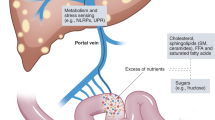
Lang, S. & Schnabl, B. Microbiota and fatty liver disease-the known, the unknown, and the future. Cell Host Microbe 28, 233–244 (2020).
Philips, C. A. et al. Healthy donor fecal microbiota transplantation in steroid-ineligible severe alcoholic hepatitis: a pilot study. Clin. Gastroenterol. Hepatol. 15, 600–602 (2017).
Leclercq, S. et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl Acad. Sci. USA 111, E4485–E4493 (2014).
Grander, C. et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 67, 891–901 (2018).
Lang, S. et al. Changes in the fecal bacterial microbiota associated with disease severity in alcoholic hepatitis patients. Gut Microbes 12, 1785251 (2020).
Mutlu, E. A. et al. Colonic microbiome is altered in alcoholism. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G966–G978 (2012).
Duan, Y. et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511 (2019).
Bode, J. C. et al. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology 31, 30–34 (1984).
Wang, L. et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe 19, 227–239 (2016).
Yang, A. M. et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Investig 127, 2829–2841 (2017).
Chu, H. et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J. Hepatol. 72, 391–400 (2020).
Jiang, L. et al. Intestinal virome in patients with alcoholic hepatitis. Hepatology 2020. https://doi.org/10.1002/hep.31459. Online ahead of print.
Bluemel, S. et al. Precision medicine in alcoholic and nonalcoholic fatty liver disease via modulating the gut microbiota. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G1018–g36 (2016).
Brenner, D. A. et al. Role of gut microbiota in liver disease. J. Clin. Gastroenterol. 49 Suppl 1, S25–S27 (2015).
Brandl, K. et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J. Hepatol. 69, 396–405 (2018).
Hendrikx, T. & Schnabl, B. Antimicrobial proteins: intestinal guards to protect against liver disease. J. Gastroenterol. 54, 209–217 (2019).
Inokuchi, S. et al. Toll-like receptor 4 mediates alcohol-induced steatohepatitis through bone marrow-derived and endogenous liver cells in mice. Alcohol Clin. Exp. Res. 35, 1509–1518 (2011).
Iracheta-Vellve, A. et al. Inhibition of sterile danger signals, uric acid and ATP, prevents inflammasome activation and protects from alcoholic steatohepatitis in mice. J. Hepatol. 63, 1147–1155 (2015).
Petrasek, J. et al. Metabolic danger signals, uric acid and ATP, mediate inflammatory cross-talk between hepatocytes and immune cells in alcoholic liver disease. J. Leukoc. Biol. 98, 249–256 (2015).
Petrasek, J. et al. IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. J. Clin. Investig. 122, 3476–3489 (2012).
Lang, S. et al. Cytolysin-positive Enterococcus faecalis is not increased in patients with non-alcoholic steatohepatitis. Liver Int. 40, 860–865 (2020).
Hartmann, P. et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology 67, 2150–2166 (2018).
Couch, R. D. et al. Alcohol induced alterations to the human fecal VOC metabolome. PLoS ONE 10, e0119362 (2015).
Smirnova, E. et al. Fecal microbiome distinguishes alcohol consumption from alcoholic hepatitis but does not discriminate disease severity. Hepatology 72, 271–286 (2020).
Cresci, G. A. et al. Tributyrin supplementation protects mice from acute ethanol-induced gut injury. Alcohol Clin. Exp. Res. 38, 1489–1501 (2014).
Li, Z. et al. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology 37, 343–350 (2003).
Alferink, L. J. et al. Microbiomics, metabolomics, predicted metagenomics and hepatic steatosis in a population-based study of 1355 adults. Hepatology 2020. https://doi.org/10.1002/hep.31417. Online ahead of print.
Zhu, L. et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013).
Mouzaki, M. et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127 (2013).
Loomba, R. et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 25, 1054–62.e5 (2017).
Lang, S. et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology 159, 1839–1852 (2020).
Scorletti, E. et al. Synbiotics alter fecal microbiomes, but not liver fat or fibrosis, in a randomized trial of patients with nonalcoholic fatty liver disease. Gastroenterology 158, 1597–610.e7 (2020).
Chong, C. Y. L. et al. Randomised double-blind placebo-controlled trial of inulin with metronidazole in non-alcoholic fatty liver disease (NAFLD). Nutrients 12, 937 (2020).
Aron-Wisnewsky, J. et al. Nonalcoholic fatty liver disease: modulating gut microbiota to improve severity? Gastroenterology 158, 1881–1898 (2020).
Tilg, H. et al. The intestinal microbiota fuelling metabolic inflammation. Nat. Rev. Immunol. 20, 40–54 (2020).
Gupta, B. et al. Western diet-induced increase in colonic bile acids compromises epithelial barrier in nonalcoholic steatohepatitis. FASEB J. 34, 7089–7102 (2020).
Craven, L. et al. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability: a randomized control trial. Am. J. Gastroenterol. 115, 1055–1065 (2020).
Chu, H. et al. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut 68, 359–370 (2019).
Zhao, M. et al. TMAVA, a metabolite of intestinal microbes, is increased in plasma from patients with liver steatosis, inhibits γ-butyrobetaine hydroxylase, and exacerbates fatty liver in mice. Gastroenterology 158, 2266–81.e27 (2020).
Hoyles, L. et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018).
Goldstein, J. L. What makes a piece of art or science a masterpiece? Cell 175, 1–5 (2018).
Day, C. P. & James, O. F. Steatohepatitis: a tale of two “hits”? Gastroenterology 114, 842–845 (1998).
Fei, N. et al. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease. mBio. 11, e03263–19 (2020).
Carpino, G. et al. Increased liver localization of lipopolysaccharides in human and experimental NAFLD. Hepatology 159, 1715–1730 (2019).
Yuan, J. et al. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab. 30, 675–88.e7 (2019).
Bajaj, J. S. et al. Serum levels of metabolites produced by intestinal microbes and lipid moieties independently associated with acute on chronic liver failure and death in patients with cirrhosis. Gastroenterology (2020).
Schierwagen, R. et al. Circulating microbiome in blood of different circulatory compartments. Gut 68, 578–580 (2019).
Sookoian, S. et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut 69, 1483–1491 (2020).
Anhê, F. F. et al. Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2, 233–242 (2020).
Wei, Y. et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 69, 569–577 (2020).
Liwinski, T. et al. A disease-specific decline of the relative abundance of Bifidobacterium in patients with autoimmune hepatitis. Aliment Pharmacol Ther. 51, 1417–1428 (2020).
Lv, L. X. et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ. Microbiol. 18, 2272–2286 (2016).
Tang, R. et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 67, 534–541 (2018).
Chen, W. et al. Comprehensive analysis of serum and fecal bile acid profiles and interaction with gut microbiota in primary biliary cholangitis. Clin. Rev. Allergy Immunol. 58, 25–38 (2020).
Loomba, R. et al. The commensal microbe veillonella as a marker for response to an FGF19 analog in nonalcoholic steatohepatitis. Hepatology 2020. https://doi.org/10.1002/hep.31523. Online ahead of print.
Furukawa, M. et al. Gut dysbiosis associated with clinical prognosis of patients with primary biliary cholangitis. Hepatol. Res. 50, 840–852 (2020).
Kummen, M. et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 66, 611–619 (2017).
Rühlemann, M. C. et al. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut 66, 753–754 (2017).
Sabino, J. et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 65, 1681–1689 (2016).
Pereira, P. et al. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS ONE 12, e0182924 (2017).
Rossen, N. G. et al. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J. Crohns Colitis 9, 342–348 (2015).
Kevans, D. et al. Characterization of intestinal microbiota in ulcerative colitis patients with and without primary sclerosing cholangitis. J. Crohns Colitis 10, 330–337 (2016).
Torres, J. et al. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment Pharmacol. Ther. 43, 790–801 (2016).
Quraishi, M. N. et al. The gut-adherent microbiota of PSC-IBD is distinct to that of IBD. Gut 66, 386–388 (2017).
Iwasawa, K. et al. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut 66, 1344–1346 (2017).
Bajer, L. et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 23, 4548–4558 (2017).
Torres, J. et al. The gut microbiota, bile acids and their correlation in primary sclerosing cholangitis associated with inflammatory bowel disease. United European Gastroenterol. J. 6, 112–122 (2018).
Ruff, W. E. et al. Host-microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol 18, 521–538 (2020).
Meng, X. et al. Microbe-metabolite-host axis, two-way action in the pathogenesis and treatment of human autoimmunity. Autoimmun. Rev. 18, 455–475 (2019).
Cai, W. et al. Intestinal microbiome and permeability in patients with autoimmune hepatitis. Best. Pract. Res Clin. Gastroenterol. 31, 669–673 (2017).
Fussey, S. P. et al. Reactivity of primary biliary cirrhosis sera with Escherichia coli dihydrolipoamide acetyltransferase (E2p): characterization of the main immunogenic region. Proc. Natl Acad. Sci. USA 87, 3987–3991 (1990).
Bogdanos, D. P. et al. Primary biliary cirrhosis is characterized by IgG3 antibodies cross-reactive with the major mitochondrial autoepitope and its Lactobacillus mimic. Hepatology 42, 458–465 (2005).
Terjung, B. et al. p-ANCAs in autoimmune liver disorders recognise human beta-tubulin isotype 5 and cross-react with microbial protein FtsZ. Gut 59, 808–816 (2010).
Manfredo Vieira, S. et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359, 1156–1161 (2018).
Nakamoto, N. et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 4, 492–503 (2019).
Trivedi, P. J. et al. Intestinal CCL25 expression is increased in colitis and correlates with inflammatory activity. J. Autoimmun. 68, 98–104 (2016).
Trivedi, P. J. et al. Vascular adhesion protein-1 is elevated in primary sclerosing cholangitis, is predictive of clinical outcome and facilitates recruitment of gut-tropic lymphocytes to liver in a substrate-dependent manner. Gut 67, 1135–1145 (2018).
Henriksen, E. K. et al. Gut and liver T-cells of common clonal origin in primary sclerosing cholangitis-inflammatory bowel disease. J. Hepatol. 66, 116–122 (2017).
Moro-Sibilot, L. et al. Mouse and human liver contain immunoglobulin A-secreting cells originating from Peyer’s patches and directed against intestinal antigens. Gastroenterology 151, 311–323 (2016).
Bajaj, J. S. & Khoruts, A. Microbiota changes and intestinal microbiota transplantation in liver diseases and cirrhosis. J. Hepatol. 72, 1003–1027 (2020).
Schwabe, R. F. & Greten, T. F. Gut microbiome in HCC—mechanisms, diagnosis and therapy. J. Hepatol. 72, 230–238 (2020).
Sehgal, R. et al. Role of microbiota in pathogenesis and management of viral hepatitis. Front. Cell Infect. Microbiol. 10, 341 (2020).
Lu, H. et al. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb Ecol. 61, 693–703 (2011).
Cui, L. et al. The human mycobiome in health and disease. Genome Med. 5, 63 (2013).
Xu, M. et al. Changes of fecal Bifidobacterium species in adult patients with hepatitis B virus-induced chronic liver disease. Microb. Ecol. 63, 304–313 (2012).
Chou, H. H. et al. Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc. Natl Acad. Sci. USA 112, 2175–2180 (2015).
Chauhan, A. et al. Fecal microbiota transplantation in hepatitis B e antigen-positive chronic hepatitis B patients: a pilot study. Dig. Dis. Sci. 2020. https://doi.org/10.1007/s10620-020-06246-x. Online ahead of print.
Aly, A. M. et al. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 8, 42 (2016).
Heidrich, B. et al. Intestinal microbiota in patients with chronic hepatitis C with and without cirrhosis compared with healthy controls. Liver Int. 38, 50–58 (2018).
Inoue, T. et al. Gut dysbiosis associated with hepatitis C virus infection. Clin. Infect. Dis. 67, 869–877 (2018).
Wu, J. et al. Altered faecal microbiota on the expression of Th cells responses in the exacerbation of patients with hepatitis E infection. J. Viral Hepat. 27, 1243–1252 (2020).
Qin, N. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014).
Bajaj, J. S. et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 60, 940–947 (2014).
Bajaj, J. S. et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G675–G685 (2012).
Santiago, A. et al. Alteration of the serum microbiome composition in cirrhotic patients with ascites. Sci. Rep. 6, 25001 (2016).
Bajaj, J. S. et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology 62, 1260–1271 (2015).
Bajaj, J. S. et al. Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin. Gastroenterol. Hepatol. 17, 756–65.e3 (2019).
Albillos, A. et al. Cirrhosis-associated immune dysfunction: distinctive features and clinical relevance. J. Hepatol. 61, 1385–1396 (2014).
Muñoz, L. et al. Intestinal immune dysregulation driven by dysbiosis promotes barrier disruption and bacterial translocation in rats with cirrhosis. Hepatology 70, 925–938 (2019).
Lorenzo-Zúñiga, V. et al. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology 37, 551–557 (2003).
Sorribas, M. et al. FXR modulates the gut-vascular barrier by regulating the entry sites for bacterial translocation in experimental cirrhosis. J. Hepatol. 71, 1126–1140 (2019).
Oh, T. G. et al. A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab. (2020).
Tilg, H. et al. Gut microbiome and liver diseases. Gut 65, 2035–2044 (2016).
Ponziani, F. R. et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 69, 107–120 (2019).
Ren, Z. et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 68, 1014–1023 (2019).
Lin, R. S. et al. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J. Hepatol. 22, 165–172 (1995).
Bellot, P. et al. Bacterial DNA translocation is associated with systemic circulatory abnormalities and intrahepatic endothelial dysfunction in patients with cirrhosis. Hepatology 52, 2044–2052 (2010).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Lagier, J. C. et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol 1, 16203 (2016).
Metwaly, A. & Haller, D. Multi-omics in IBD biomarker discovery: the missing links. Nat. Rev. Gastroenterol. Hepatol. 16, 587–588 (2019).
Zhang, Q. et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut 68, 2019–2031 (2019).
Burberry, A. et al. C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature 582, 89–94 (2020).
Hu, S. et al. Whole exome sequencing analyses reveal gene-microbiota interactions in the context of IBD. Gut 2020;gutjnl-2019-319706. https://doi.org/10.1136/gutjnl-2019-319706. Online ahead of print.
Bober, J. R. et al. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu. Rev. Biomed. Eng. 20, 277–300 (2018).
Hendrikx, T. et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515 (2019).
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (grants #81830016, 81771732, and 81620108002 to X.M.; #81922010 and 81873561 to R.T.). This study was supported in part by services provided by the NIH centers P30 DK120515 and P50 AA011999. H.T. is supported by the excellence initiative VASCage (Centre for Promoting Vascular Health in the Ageing Community), an R&D K-Centre (COMET program—Competence Centers for Excellent Technologies) funded by the Austrian Ministry for Transport, Innovation and Technology, the Austrian Ministry for Digital and Economic Affairs and the federal states Tyrol, Salzburg and Vienna.
Author information
Authors and Affiliations
Contributions
B.S. was responsible for writing the chapter “Contribution of the gut microbiota to alcohol-associated liver disease”. H.T. wrote the chapter “NAFLD and the Microbiome”. R.W., R.T. and X.M. wrote the chapters “The Gut Microbiome”, “The Gut Microbiome and Liver Immunology”, “The Gut Microbiome and AILD”, and “The Gut Microbiome and Other Types of Liver Diseases”. B.L. was responsible for draft calibration.
Corresponding authors
Ethics declarations
Competing interests
B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics, Mabwell Therapeutics and Patara Pharmaceuticals. B.S.’s institution (UC San Diego) has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company and Axial Biotherapeutics. H.T. and X.M. have no competing interests.
Rights and permissions
About this article
Cite this article
Wang, R., Tang, R., Li, B. et al. Gut microbiome, liver immunology, and liver diseases. Cell Mol Immunol 18, 4–17 (2021). https://doi.org/10.1038/s41423-020-00592-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-020-00592-6
Keywords
This article is cited by
-
Large-scale causal analysis of gut microbiota and six common complications of diabetes: a mendelian randomization study
Diabetology & Metabolic Syndrome (2024)
-
Impact of perinatal administration of probiotics on immune cell composition in neonatal mice
Pediatric Research (2024)
-
Immunology of gut microbiome and liver in non-alcoholic fatty liver disease (NAFLD): mechanisms, bacteria, and novel therapeutic targets
Archives of Microbiology (2024)
-
Elevated plasma and bile levels of corisin, a microbiota-derived proapoptotic peptide, in patients with severe acute cholangitis
Gut Pathogens (2023)
-
Metagenomic sequencing reveals altered gut microbial compositions and gene functions in patients with non-segmental vitiligo
BMC Microbiology (2023)