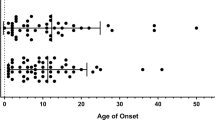
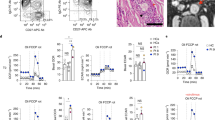
Abstract
Predominantly antibody deficiency (PAD) is the most prevalent form of primary immunodeficiency, and is characterized by broad clinical, immunological and genetic heterogeneity. Utilizing the current gold standard of whole exome sequencing for diagnosis, pathogenic gene variants are only identified in less than 20% of patients. While elucidation of the causal genes underlying PAD has provided many insights into the cellular and molecular mechanisms underpinning disease pathogenesis, many other genes may remain as yet undefined to enable definitive diagnosis, prognostic monitoring and targeted therapy of patients. Considering that many patients display a relatively late onset of disease presentation in their 2nd or 3rd decade of life, it is questionable whether a single genetic lesion underlies disease in all patients. Potentially, combined effects of other gene variants and/or non-genetic factors, including specific infections can drive disease presentation. In this review, we define (1) the clinical and immunological variability of PAD, (2) consider how genetic defects identified in PAD have given insight into B-cell immunobiology, (3) address recent technological advances in genomics and the challenges associated with identifying causal variants, and (4) discuss how functional validation of variants of unknown significance could potentially be translated into increased diagnostic rates, improved prognostic monitoring and personalized medicine for PAD patients. A multidisciplinary approach will be the key to curtailing the early mortality and high morbidity rates in this immune disorder.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kobrynski, L., Powell, R. W. & Bowen, S. Prevalence and morbidity of primary immunodeficiency diseases, United States 2001–2007. J. Clin. Immunol. 34, 954–961 (2014).
Picard, C. & Fischer, A. Contribution of high-throughput DNA sequencing to the study of primary immunodeficiencies. Eur. J. Immunol. 44, 2854–2861 (2014).
Bucciol, G. et al. Lessons learned from the study of human inborn errors of innate immunity. J. Allergy Clin. Immunol. 143, 507–527 (2019).
Tangye, S. G. et al. Human inborn errors of immunity: 2019 update on the classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 40, 66–81 (2020).
Bousfiha, A. et al. Human inborn errors of immunity: 2019 update of the IUIS phenotypical classification. J. Clin. Immunol. 40, 66–81 (2020).
Gathmann, B. et al. The European internet-based patient and research database for primary immunodeficiencies: results 2006-2008. Clin. Exp. Immunol. 157(Suppl 1), 3–11 (2009).
Durandy, A., Kracker, S. & Fischer, A. Primary antibody deficiencies. Nat. Rev. Immunol. 13, 519–533 (2013).
Lucas, M. et al. Infection outcomes in patients with common variable immunodeficiency disorders: relationship to immunoglobulin therapy over 22 years. J. Allergy Clin. Immunol. 125, 1354–1360.e1354 (2010).
Bogaert, D. J. et al. Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J. Med. Genet. 53, 575–590 (2016).
Aggarwal, V., Banday, A. Z., Jindal, A. K., Das, J. & Rawat, A. Recent advances in elucidating the genetics of common variable immunodeficiency. Genes Dis. 7, 26–37 (2020).
Bonilla, F. A. et al. International Consensus Document (ICON): common variable immunodeficiency disorders. J. Allergy Clin. Immunol. Pract. 4, 38–59 (2016).
Tsukada, S. et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell 72, 279–290 (1993).
Vetrie, D. et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature 361, 226–233 (1993).
Vorechovsky, I. et al. Molecular diagnosis of X-linked agammaglobulinaemia. Lancet 341, 1153 (1993).
Dingjan, G. M. et al. Bruton’s tyrosine kinase regulates the activation of gene rearrangements at the lambda light chain locus in precursor B cells in the mouse. J. Exp. Med. 193, 1169–1178 (2001).
Papapietro, O. et al. Topoisomerase 2beta mutation impairs early B cell development. Blood 135, 1497–1501 (2020).
Ben-Ali, M. et al. Homozygous transcription factor 3 gene (TCF3) mutation is associated with severe hypogammaglobulinemia and B-cell acute lymphoblastic leukemia. J. Allergy Clin. Immunol. 140, 1191–1194.e1194 (2017).
Qureshi, S., Sheikh, M. D. A. & Qamar, F. N. Autosomal recessive agammaglobulinemia—first case with a novel TCF3 mutation from Pakistan. Clin. Immunol. 198, 100–101 (2019).
Kuehn, H. S. et al. Loss of B Cells in Patients with Heterozygous Mutations in IKAROS. N. Engl. J. Med. 374, 1032–1043 (2016).
Yel, L. et al. Mutations in the mu heavy-chain gene in patients with agammaglobulinemia. N. Engl. J. Med. 335, 1486–1493 (1996).
Minegishi, Y. et al. Mutations in Igalpha (CD79a) result in a complete block in B-cell development. J. Clin. Investig. 104, 1115–1121 (1999).
Minegishi, Y. et al. Mutations in the human lambda5/14.1 gene result in B cell deficiency and agammaglobulinemia. J. Exp. Med. 187, 71–77 (1998).
Minegishi, Y. et al. An essential role for BLNK in human B cell development. Science 286, 1954–1957 (1999).
Dobbs, A. K. et al. Cutting edge: a hypomorphic mutation in Igbeta (CD79b) in a patient with immunodeficiency and a leaky defect in B cell development. J. Immunol. 179, 2055–2059 (2007).
Anzilotti, C. et al. An essential role for the Zn(2+) transporter ZIP7 in B cell development. Nat. Immunol. 20, 350–361 (2019).
Broderick, L. et al. Mutations in topoisomerase IIbeta result in a B cell immunodeficiency. Nat. Commun. 10, 3644 (2019).
Croker, B. A. et al. The Rac2 guanosine triphosphatase regulates B lymphocyte antigen receptor responses and chemotaxis and is required for establishment of B-1a and marginal zone B lymphocytes. J. Immunol. 168, 3376–3386 (2002).
Alkhairy, O. K. et al. RAC2 loss-of-function mutation in 2 siblings with characteristics of common variable immunodeficiency. J. Allergy Clin. Immunol. 135, 1380–1384.e1-5 (2015).
Elkaim, E. et al. Clinical and immunologic phenotype associated with activated phosphoinositide 3-kinase delta syndrome 2: a cohort study. J. Allergy Clin. Immunol. 138, 210–218.e219 (2016).
Sogkas, G. et al. Primary immunodeficiency disorder caused by phosphoinositide 3-kinase delta deficiency. J. Allergy Clin. Immunol. 142, 1650–1653 e1652 (2018).
Cohen, S. B. et al. Human primary immunodeficiency caused by expression of a kinase-dead p110delta mutant. J. Allergy Clin. Immunol. 143, 797–799 e792 (2019).
Ramadani, F. et al. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci. Signal. 3, ra60 (2010).
Conley, M. E. et al. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85alpha subunit of PI3K. J. Exp. Med 209, 463–470 (2012).
Tang, P. et al. Autosomal recessive agammaglobulinemia due to a homozygous mutation in PIK3R1. J. Clin. Immunol. 38, 88–95 (2018).
Ferrari, S. et al. Mutations of the Igbeta gene cause agammaglobulinemia in man. J. Exp. Med. 204, 2047–2051 (2007).
van Zelm, M. C. et al. Gross deletions involving IGHM, BTK, or Artemis: a model for genomic lesions mediated by transposable elements. Am. J. Hum. Genet. 82, 320–332 (2008).
Ameratunga, R. et al. Comparison of diagnostic criteria for common variable immunodeficiency disorder. Front. Immunol. 5, 415 (2014).
Boileau, J. et al. Autoimmunity in common variable immunodeficiency: correlation with lymphocyte phenotype in the French DEFI study. J. Autoimmun. 36, 25–32 (2011).
Immunodeficiencies, E. S. F. New Clinical Diagnosis Criteria for the ESID Registry. Geneva. https://esid.org/Working-Parties/Registry/Diagnosis-criteria (2017).
Conley, M. E., Notarangelo, L. D. & Etzioni, A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies). Clin. Immunol. 93, 190–197 (1999).
von Spee-Mayer, C. et al. Evaluating laboratory criteria for combined immunodeficiency in adult patients diagnosed with common variable immunodeficiency. Clin. Immunol. 203, 59–62 (2019).
Bertinchamp, R. et al. Exclusion of patients with a severe T-cell defect improves the definition of common variable immunodeficiency. J. Allergy Clin. Immunol. Pract. 4, 1147–1157 (2016).
Chapel, H. Common variable immunodeficiency disorders (CVID)—diagnoses of exclusion, especially combined immune defects. J. Allergy Clin. Immunol. Pr. 4, 1158–1159 (2016).
Grosserichter-Wagener, C. et al. Defective formation of IgA memory B cells, Th1 and Th17 cells in symptomatic patients with selective IgA deficiency. Clin. Transl. Immunol. 9, e1130 (2020).
Driessen, G. J. et al. Common variable immunodeficiency and idiopathic primary hypogammaglobulinemia: two different conditions within the same disease spectrum. Haematologica 98, 1617–1623 (2013).
Cheng, Y. K., Decker, P. A., O’Byrne, M. M. & Weiler, C. R. Specific polysaccharide antibody deficiency syndrome (SPAD): Clinical and laboratory characteristics of seventy seven patients. J. Allergy Clin. Immunol. 115, S158 (2005).
Kim, J. H. et al. Immunoglobulin G subclass deficiency is the major phenotype of primary immunodeficiency in a Korean adult cohort. J. Korean Med. Sci. 25, 824–828 (2010).
Resnick, E. S., Moshier, E. L., Godbold, J. H. & Cunningham-Rundles, C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood 119, 1650–1657 (2012).
Jolles, S. Subclinical infection and dosing in primary immunodeficiencies. Clin. Exp. Immunol. 178(Suppl 1), 67–69 (2014).
Edwards, E. S. J. et al. Predominantly antibody-deficient patients with non-infectious complications have reduced naive B, Treg, Th17, and Tfh17 cells. Front. Immunol. 10, 2593 (2019).
Slade, C. A. et al. Delayed diagnosis and complications of predominantly antibody deficiencies in a cohort of australian adults. Front. Immunol. 9, 694 (2018).
Chapel, H. et al. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood 112, 277–286 (2008).
Odnoletkova, I. et al. The burden of common variable immunodeficiency disorders: a retrospective analysis of the European Society for Immunodeficiency (ESID) registry data. Orphanet J. Rare Dis. 13, 201 (2018).
Romberg, N. et al. Patients with common variable immunodeficiency with autoimmune cytopenias exhibit hyperplastic yet inefficient germinal center responses. J. Allergy Clin. Immunol. 143, 258–265 (2019).
Warnatz, K. et al. Severe deficiency of switched memory B cells (CD27(+)IgM(-)IgD(-)) in subgroups of patients with common variable immunodeficiency: a new approach to classify a heterogeneous disease. Blood 99, 1544–1551 (2002).
Wehr, C. et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood 111, 77–85 (2008).
Blanco, E. et al. Defects in memory B-cell and plasma cell subsets expressing different immunoglobulin-subclasses in patients with CVID and immunoglobulin subclass deficiencies. J. Allergy Clin. Immunol. 144, 809–824 (2019).
Driessen, G. J. et al. B-cell replication history and somatic hypermutation status identify distinct pathophysiologic backgrounds in common variable immunodeficiency. Blood 118, 6814–6823 (2011).
Warnatz, K. & Schlesier, M. Flowcytometric phenotyping of common variable immunodeficiency. Cytom. B Clin. Cytom. 74, 261–271 (2008).
Ebbo, M. et al. Low circulating natural killer cell counts are associated with severe disease in patients with common variable immunodeficiency. EBio Med. 6, 222–230 (2016).
Aspalter, R. M., Sewell, W. A., Dolman, K., Farrant, J. & Webster, A. D. Deficiency in circulating natural killer (NK) cell subsets in common variable immunodeficiency and X-linked agammaglobulinaemia. Clin. Exp. Immunol. 121, 506–514 (2000).
Giovannetti, A. et al. Unravelling the complexity of T cell abnormalities in common variable immunodeficiency. J. Immunol. 178, 3932–3943 (2007).
Bateman, E. A. et al. T cell phenotypes in patients with common variable immunodeficiency disorders: associations with clinical phenotypes in comparison with other groups with recurrent infections. Clin. Exp. Immunol. 170, 202–211 (2012).
Coraglia, A. et al. Common variable immunodeficiency and circulating TFH. J. Immunol. Res. 2016, 4951587 (2016).
Unger, S. et al. The TH1 phenotype of follicular helper T cells indicates an IFN-gamma-associated immune dysregulation in patients with CD21low common variable immunodeficiency. J. Allergy Clin. Immunol. 141, 730–740 (2018).
Romberg, N. D., Hsu, I., Price, C. C., Cunningham-Rundles, C. & Meffre, E. Expansion of circulating T follicular helper cells in CVID patients with autoimmune cytopenias. J. Allergy Clin. Immunol. 133, AB162 (2014).
Barbosa, R. R. et al. Primary B-cell deficiencies reveal a link between human IL-17-producing CD4 T-cell homeostasis and B-cell differentiation. PLoS ONE 6, e22848 (2011).
Melo, K. M. et al. A decreased frequency of regulatory T cells in patients with common variable immunodeficiency. PLoS ONE 4, e6269 (2009).
Arumugakani, G., Wood, P. M. & Carter, C. R. Frequency of Treg cells is reduced in CVID patients with autoimmunity and splenomegaly and is associated with expanded CD21lo B lymphocytes. J. Clin. Immunol. 30, 292–300 (2010).
Malphettes, M. et al. Late-onset combined immune deficiency: a subset of common variable immunodeficiency with severe T cell defect. Clin. Infect. Dis. 49, 1329–1338 (2009).
Hultberg, J., Ernerudh, J., Larsson, M., Nilsdotter-Augustinsson, A. & Nystrom, S. Plasma protein profiling reflects TH1-driven immune dysregulation in common variable immunodeficiency. J. Allergy Clin. Immunol. 146, 417–428 (2020).
Cols, M. et al. Expansion of inflammatory innate lymphoid cells in patients with common variable immune deficiency. J. Allergy Clin. Immunol. 137, 1206–1215 e1206 (2016).
Berkowska, M. A., van der Burg, M., van Dongen, J. J. & van Zelm, M. C. Checkpoints of B cell differentiation: visualizing Ig-centric processes. Ann. N.Y. Acad. Sci. 1246, 11–25 (2011).
Clark, M. R., Mandal, M., Ochiai, K. & Singh, H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 14, 69–80 (2014).
Boisson, B. et al. A recurrent dominant negative E47 mutation causes agammaglobulinemia and BCR(-) B cells. J. Clin. Investig. 123, 4781–4785 (2013).
Rohrer, J., Minegishi, Y., Richter, D., Eguiguren, J. & Conley, M. E. Unusual mutations in Btk: an insertion, a duplication, an inversion, and four large deletions. Clin. Immunol. 90, 28–37 (1999).
Vihinen, M. et al. Mutations of the human BTK gene coding for bruton tyrosine kinase in X-linked agammaglobulinemia. Hum. Mutat. 13, 280–285 (1999).
Kumaki, E. et al. Atypical SIFD with novel TRNT1 mutations: a case study on the pathogenesis of B-cell deficiency. Int. J. Hematol. 109, 382–389 (2019).
Wedatilake, Y. et al. TRNT1 deficiency: clinical, biochemical and molecular genetic features. Orphanet J. Rare Dis. 11, 90 (2016).
Meyers, G. et al. Activation-induced cytidine deaminase (AID) is required for B-cell tolerance in humans. Proc. Natl Acad. Sci. USA 108, 11554–11559 (2011).
Warnatz, K. et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc. Natl Acad. Sci. USA 106, 13945–13950 (2009).
Losi, C. G. et al. Mutational analysis of human BAFF receptor TNFRSF13C (BAFF-R) in patients with common variable immunodeficiency. J. Clin. Immunol. 25, 496–502 (2005).
Wang, H. Y. et al. Antibody deficiency associated with an inherited autosomal dominant mutation in TWEAK. Proc. Natl Acad. Sci. USA 110, 5127–5132 (2013).
Cherukuri, A., Cheng, P. C., Sohn, H. W. & Pierce, S. K. The CD19/CD21 complex functions to prolong B cell antigen receptor signaling from lipid rafts. Immunity 14, 169–179 (2001).
Carter, R. H. & Fearon, D. T. CD19: lowering the threshold for antigen receptor stimulation of B lymphocytes. Science 256, 105–107 (1992).
van Noesel, C. J., Lankester, A. C. & van Lier, R. A. Dual antigen recognition by B cells. Immunol. Today 14, 8–11 (1993).
van Zelm, M. C. et al. Human CD19 and CD40L deficiencies impair antibody selection and differentially affect somatic hypermutation. J. Allergy Clin. Immunol. 134, 135–144 (2014).
van Zelm, M. C. et al. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Investig. 120, 1265–1274 (2010).
van Zelm, M. C. et al. Antibody deficiency due to a missense mutation in CD19 demonstrates the importance of the conserved tryptophan 41 in immunoglobulin superfamily domain formation. Hum. Mol. Genet. 20, 1854–1863 (2011).
Wentink, M. W. et al. CD21 and CD19 deficiency: two defects in the same complex leading to different disease modalities. Clin. Immunol. 161, 120–127 (2015).
Thiel, J. et al. Genetic CD21 deficiency is associated with hypogammaglobulinemia. J. Allergy Clin. Immunol. 129, 801–810 e806 (2012).
Jansen, E. J. et al. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat. Commun. 7, 11600 (2016).
Oellerich, T. et al. The B-cell antigen receptor signals through a preformed transducer module of SLP65 and CIN85. EMBO J. 30, 3620–3634 (2011).
Keller, B. et al. Germline deletion of CIN85 in humans with X chromosome-linked antibody deficiency. J. Exp. Med. 215, 1327–1336 (2018).
van Zelm, M. C. et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. N. Engl. J. Med. 354, 1901–1912 (2006).
Kuijpers, T. W. et al. CD20 deficiency in humans results in impaired T cell-independent antibody responses. J. Clin. Investig. 120, 214–222 (2010).
Snow, A. L. et al. Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J. Exp. Med. 209, 2247–2261 (2012).
Polyak, M. J., Li, H., Shariat, N. & Deans, J. P. CD20 homo-oligomers physically associate with the B cell antigen receptor. Dissociation upon receptor engagement and recruitment of phosphoproteins and calmodulin-binding proteins. J. Biol. Chem. 283, 18545–18552 (2008).
Petrie, R. J. & Deans, J. P. Colocalization of the B cell receptor and CD20 followed by activation-dependent dissociation in distinct lipid rafts. J. Immunol. 169, 2886–2891 (2002).
Kranich, J. & Krautler, N. J. How follicular dendritic cells shape the B-Cell antigenome. Front. Immunol. 7, 225 (2016).
MacLennan, I. C. Germinal centers. Annu. Rev. Immunol. 12, 117–139 (1994).
Bhushan, A. & Covey, L. R. CD40:CD40L interactions in X-linked and non-X-linked hyper-IgM syndromes. Immunol. Res. 24, 311–324 (2001).
Castigli, E. et al. CD40 ligand/CD40 deficiency. Int. Arch. Allergy Immunol. 107, 37–39 (1995).
Chou, J. et al. A novel mutation in ICOS presenting as hypogammaglobulinemia with susceptibility to opportunistic pathogens. J. Allergy Clin. Immunol. 136, 794–797.e791 (2015).
Grimbacher, B. et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat. Immunol. 4, 261–268 (2003).
Salzer, U. et al. ICOS deficiency in patients with common variable immunodeficiency. Clin. Immunol. 113, 234–240 (2004).
Warnatz, K. et al. Human ICOS deficiency abrogates the germinal center reaction and provides a monogenic model for common variable immunodeficiency. Blood 107, 3045–3052 (2006).
Roussel, L. et al. Loss of human ICOSL results in combined immunodeficiency. J. Exp. Med. 215, 3151–3164 (2018).
Revy, P. et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102, 565–575 (2000).
Imai, K. et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4, 1023–1028 (2003).
Gardes, P. et al. Human MSH6 deficiency is associated with impaired antibody maturation. J. Immunol. 188, 2023–2029 (2012).
Kracker, S. et al. An inherited immunoglobulin class-switch recombination deficiency associated with a defect in the INO80 chromatin remodeling complex. J. Allergy Clin. Immunol. 135, 998–1007 e1006 (2015).
Castigli, E. et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat. Genet. 37, 829–834 (2005).
Knight, A. K. et al. High serum levels of BAFF, APRIL, and TACI in common variable immunodeficiency. Clin. Immunol. 124, 182–189 (2007).
Pan-Hammarstrom, Q. et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat. Genet. 39, 429–430 (2007).
Romberg, N. et al. CVID-associated TACI mutations affect autoreactive B cell selection and activation. J. Clin. Investig. 123, 4283–4293 (2013).
Salzer, U. et al. Relevance of biallelic versus monoallelic TNFRSF13B mutations in distinguishing disease-causing from risk-increasing TNFRSF13B variants in antibody deficiency syndromes. Blood 113, 1967–1976 (2009).
Salzer, U. et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat. Genet. 37, 820–828 (2005).
Zhang, L. et al. Transmembrane activator and calcium-modulating cyclophilin ligand interactor mutations in common variable immunodeficiency: clinical and immunologic outcomes in heterozygotes. J. Allergy Clin. Immunol. 120, 1178–1185 (2007).
Almejun, M. B. et al. Naturally occurring mutation affecting the MyD88-binding site of TNFRSF13B impairs triggering of class switch recombination. Eur. J. Immunol. 43, 805–814 (2013).
Scott, O. & Roifman, C. M. NF-kappaB pathway and the Goldilocks principle: Lessons from human disorders of immunity and inflammation. J. Allergy Clin. Immunol. 143, 1688–1701 (2019).
Tangye, S. G. et al. Immune dysregulation and disease pathogenesis due to activating mutations in PIK3CD-the Goldilocks’ effect. J. Clin. Immunol. 39, 148–158 (2019).
Deau, M. C. et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J. Clin. Investig. 124, 3923–3928 (2014).
Lucas, C. L. et al. Heterozygous splice mutation in PIK3R1 causes human immunodeficiency with lymphoproliferation due to dominant activation of PI3K. J. Exp. Med. 211, 2537–2547 (2014).
Avery, D. T. et al. Germline-activating mutations in PIK3CD compromise B cell development and function. J. Exp. Med. 215, 2073–2095 (2018).
Wentink, M. et al. Genetic defects in PI3Kdelta affect B-cell differentiation and maturation leading to hypogammaglobulineamia and recurrent infections. Clin. Immunol. 176, 77–86 (2017).
Bouafia, A. et al. Loss of ARHGEF1 causes a human primary antibody deficiency. J. Clin. Investig. 129, 1047–1060 (2019).
Mathew, D., Kremer, K. N. & Torres, R. M. ARHGEF1 deficiency reveals Galpha13-associated GPCRs are critical regulators of human lymphocyte function. J. Clin. Investig. 129, 965–968 (2019).
Tsujita, Y. et al. Phosphatase and tensin homolog (PTEN) mutation can cause activated phosphatidylinositol 3-kinase delta syndrome-like immunodeficiency. J. Allergy Clin. Immunol. 138, 1672–1680 e1610 (2016).
Browning, M. J., Chandra, A., Carbonaro, V., Okkenhaug, K. & Barwell, J. Cowden’s syndrome with immunodeficiency. J. Med. Genet. 52, 856–859 (2015).
Dornan, G. L. et al. Conformational disruption of PI3Kdelta regulation by immunodeficiency mutations in PIK3CD and PIK3R1. Proc. Natl Acad. Sci. USA 114, 1982–1987 (2017).
Kaustio, M. et al. Damaging heterozygous mutations in NFKB1 lead to diverse immunologic phenotypes. J. Allergy Clin. Immunol. 140, 782–796 (2017).
Lorenzini, T. et al. Characterization of the clinical and immunological phenotype and management of 157 individuals with 56 distinct heterozygous NFKB1 mutations. J Allergy Clin. Immunol. (2020). (In press)
Tuijnenburg, P. et al. Loss-of-function nuclear factor kappaB subunit 1 (NFKB1) variants are the most common monogenic cause of common variable immunodeficiency in Europeans. J. Allergy Clin. Immunol. 142, 1285–1296 (2018).
Kuehn, H. S. et al. Novel nonsense gain-of-function NFKB2 mutations associated with a combined immunodeficiency phenotype. Blood 130, 1553–1564 (2017).
Boztug, H. et al. NF-kappaB1 haploinsufficiency causing immunodeficiency and ebv-driven lymphoproliferation. J. Clin. Immunol. 36, 533–540 (2016).
Fliegauf, M. et al. Haploinsufficiency of the NF-kappaB1 subunit p50 in common variable immunodeficiency. Am. J. Hum. Genet. 97, 389–403 (2015).
Aird, A. et al. Novel heterozygous mutation in NFKB2 is associated with early onset CVID and a functional defect in NK cells complicated by disseminated CMV infection and severe nephrotic syndrome. Front. Pediatr. 7, 303 (2019).
Brue, T. et al. Mutations in NFKB2 and potential genetic heterogeneity in patients with DAVID syndrome, having variable endocrine and immune deficiencies. BMC Med. Genet. 15, 139 (2014).
Chen, K. et al. Germline mutations in NFKB2 implicate the noncanonical NF-kappaB pathway in the pathogenesis of common variable immunodeficiency. Am. J. Hum. Genet. 93, 812–824 (2013).
Klemann, C. et al. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front. Immunol. 10, 297 (2019).
Lee, C. E. et al. Autosomal-dominant B-cell deficiency with alopecia due to a mutation in NFKB2 that results in nonprocessable p100. Blood 124, 2964–2972 (2014).
Lindsley, A. W. et al. Combined immune deficiency in a patient with a novel NFKB2 mutation. J. Clin. Immunol. 34, 910–915 (2014).
Shi, C. et al. NFKB2 mutation in common variable immunodeficiency and isolated adrenocorticotropic hormone deficiency: a case report and review of literature. Medicine 95, e5081 (2016).
Angulo, I. et al. Phosphoinositide 3-kinase delta gene mutation predisposes to respiratory infection and airway damage. Science 342, 866–871 (2013).
Coulter, T. I. et al. Clinical spectrum and features of activated phosphoinositide 3-kinase delta syndrome: a large patient cohort study. J. Allergy Clin. Immunol. 139, 597–606.e594 (2017).
Lucas, C. L., Chandra, A., Nejentsev, S., Condliffe, A. M. & Okkenhaug, K. PI3Kdelta and primary immunodeficiencies. Nat. Rev. Immunol. 16, 702–714 (2016).
Lucas, C. L. et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nat. Immunol. 15, 88–97 (2014).
Preite, S., Gomez-Rodriguez, J., Cannons, J. L. & Schwartzberg, P. L. T and B-cell signaling in activated PI3K delta syndrome: from immunodeficiency to autoimmunity. Immunol. Rev. 291, 154–173 (2019).
Bier, J. et al. Activating mutations in PIK3CD disrupt the differentiation and function of human and murine CD4(+) T cells. J. Allergy Clin. Immunol. 144, 236–253 (2019).
Edwards, E. S. J. et al. Activating PIK3CD mutations impair human cytotoxic lymphocyte differentiation and function and EBV immunity. J. Allergy Clin. Immunol. 143, 276–291.e276 (2019).
Ruiz-Garcia, R. et al. Mutations in PI3K110delta cause impaired natural killer cell function partially rescued by rapamycin treatment. J. Allergy Clin. Immunol. 142, 605–617.e607 (2018).
Perez-Andres, M. et al. Human peripheral blood B-cell compartments: a crossroad in B-cell traffic. Cytom. B Clin. Cytom. 78(Suppl 1), S47–S60 (2010).
Bolar, N. A. et al. Heterozygous loss-of-function SEC61A1 mutations cause autosomal-dominant tubulo-interstitial and glomerulocystic kidney disease with anemia. Am. J. Hum. Genet. 99, 174–187 (2016).
Schubert, D. et al. Plasma cell deficiency in human subjects with heterozygous mutations in Sec61 translocon alpha 1 subunit (SEC61A1). J. Allergy Clin. Immunol. 141, 1427–1438 (2018).
Keller, M. D. et al. Mutation in IRF2BP2 is responsible for a familial form of common variable immunodeficiency disorder. J. Allergy Clin. Immunol. 138, 544–550 e544 (2016).
Yeh, T. W. O., et al. APRIL-dependent life-long plasmacyte maintenance and immunoglobulin production in humans. J. Allergy Clin. Immunol. S0091-6749(20)30432-2 (2020). [Epub ahead of print].
Sadat, M. A. et al. Glycosylation, hypogammaglobulinemia, and resistance to viral infections. N. Engl. J. Med. 370, 1615–1625 (2014).
Bruton, O. C. Agammaglobulinemia. Pediatrics 9, 722–728 (1952).
Chinn, I. K. et al. Diagnostic interpretation of genetic studies in patients with primary immunodeficiency diseases: a working group report of the Primary Immunodeficiency Diseases Committee of the American Academy of Allergy, Asthma & Immunology. J. Allergy Clin. Immunol. 145, 46–69 (2020).
Meyts, I. et al. Exome and genome sequencing for inborn errors of immunity. J. Allergy Clin. Immunol. 138, 957–969 (2016).
Itan, Y. & Casanova, J. L. Novel primary immunodeficiency candidate genes predicted by the human gene connectome. Front. Immunol. 6, 142 (2015).
Mu, W., Lu, H. M., Chen, J., Li, S. & Elliott, A. M. Sanger confirmation is required to achieve optimal sensitivity and specificity in next-generation sequencing panel testing. J. Mol. Diagn. 18, 923–932 (2016).
Collins, F. S., Morgan, M. & Patrinos, A. The Human Genome Project: lessons from large-scale biology. Science 300, 286–290 (2003).
International Human Genome Sequencing, C. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Fang, M., Abolhassani, H., Lim, C. K., Zhang, J. & Hammarstrom, L. Next generation sequencing data analysis in primary immunodeficiency disorders—future directions. J. Clin. Immunol. 36(Suppl 1), 68–75 (2016).
Conley, M. E. & Casanova, J. L. Discovery of single-gene inborn errors of immunity by next generation sequencing. Curr. Opin. Immunol. 30, 17–23 (2014).
Oliveira, J. B. & Fleisher, T. A. Laboratory evaluation of primary immunodeficiencies. J. Allergy Clin. Immunol. 125, S297–S305 (2010).
Moens, L. N. et al. Diagnostics of primary immunodeficiency diseases: a sequencing capture approach. PLoS ONE 9, e114901 (2014).
Nijman, I. J. et al. Targeted next-generation sequencing: a novel diagnostic tool for primary immunodeficiencies. J. Allergy Clin. Immunol. 133, 529–534 (2014).
Stoddard, J. L., Niemela, J. E., Fleisher, T. A. & Rosenzweig, S. D. Targeted NGS: a cost-effective approach to molecular diagnosis of PIDs. Front. Immunol. 5, 531 (2014).
Chou, J., Ohsumi, T. K. & Geha, R. S. Use of whole exome and genome sequencing in the identification of genetic causes of primary immunodeficiencies. Curr. Opin. Allergy Clin. Immunol. 12, 623–628 (2012).
Bamshad, M. J. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755 (2011).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 39, 1522–1527 (2007).
Dyer, L. et al. Gene dosage defects in primary immunodeficiencies and related disorders: a pilot study. J. Transl. Genet. Genom. 1, 23–27 (2017).
Thaventhiran, J. E. D. et al. Whole-genome sequencing of a sporadic primary immunodeficiency cohort. Nature 583, 90–95 (2020).
Belkadi, A. et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl Acad. Sci. USA 112, 5473–5478 (2015).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Stenson, P. D. et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133, 1–9 (2014).
Jalali Sefid Dashti, M. & Gamieldien, J. A practical guide to filtering and prioritizing genetic variants. Biotechniques 62, 18–30 (2017).
Kobayashi, Y. et al. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 9, 13 (2017).
Casanova, J. L., Conley, M. E., Seligman, S. J., Abel, L. & Notarangelo, L. D. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J. Exp. Med. 211, 2137–2149 (2014).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Fischer, A. & Rausell, A. What do primary immunodeficiencies tell us about the essentiality/redundancy of immune responses? Semin. Immunol. 36, 13–16 (2018).
Zhang, S. Y. et al. Human inborn errors of immunity to infection affecting cells other than leukocytes: from the immune system to the whole organism. Curr. Opin. Immunol. 59, 88–100 (2019).
Ma, C. A. et al. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat. Genet. 49, 1192–1201 (2017).
Felgentreff, K. et al. Functional analysis of naturally occurring DCLRE1C mutations and correlation with the clinical phenotype of ARTEMIS deficiency. J. Allergy Clin. Immunol. 136, 140–150.e147 (2015).
Gutierrez-Rodrigues, F. & Calado, R. T. The interpretation of rare or novel variants: damaging vs. disease-causing. Rev. Bras. Hematol. Hemoter. 40, 3–4 (2018).
Gallo, V. et al. Diagnostics of primary immunodeficiencies through next-generation sequencing. Front. Immunol. 7, 466 (2016).
van Schouwenburg, P. A. et al. Application of whole genome and RNA sequencing to investigate the genomic landscape of common variable immunodeficiency disorders. Clin. Immunol. 160, 301–314 (2015).
de Valles-Ibanez, G. et al. Evaluating the genetics of common variable immunodeficiency: monogenetic model and beyond. Front. Immunol. 9, 636 (2018).
Christiansen, M. et al. Identification of novel genetic variants in CVID patients with autoimmunity, autoinflammation, or malignancy. Front. Immunol. 10, 3022 (2019).
Stuchly, J. et al. Common Variable Immunodeficiency patients with a phenotypic profile of immunosenescence present with thrombocytopenia. Sci. Rep. 7, 39710 (2017).
Maffucci, P. et al. Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front. Immunol. 7, 220 (2016).
Abolhassani, H. et al. Clinical implications of systematic phenotyping and exome sequencing in patients with primary antibody deficiency. Genet. Med. 21, 243–251 (2019).
Schwab, C. et al. Phenotype, penetrance, and treatment of 133 cytotoxic T-lymphocyte antigen 4-insufficient subjects. J. Allergy Clin. Immunol. 142, 1932–1946 (2018).
Mahmoud, M. et al. Structural variant calling: the long and the short of it. Genome Biol. 20, 246 (2019).
Keller, M. et al. Burden of copy number variation in common variable immunodeficiency. Clin. Exp. Immunol. 177, 269–271 (2014).
Stray-Pedersen, A. et al. Primary immunodeficiency diseases: genomic approaches delineate heterogeneous Mendelian disorders. J. Allergy Clin. Immunol. 139, 232–245 (2017).
Orange, J. S. et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J. Allergy Clin. Immunol. 127, 1360–1367.e1366 (2011).
de Bakker, P. I. et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38, 1166–1172 (2006).
Li, J. et al. Association of CLEC16A with human common variable immunodeficiency disorder and role in murine B cells. Nat. Commun. 6, 6804 (2015).
Kumar, V., Wijmenga, C. & Xavier, R. J. Genetics of immune-mediated disorders: from genome-wide association to molecular mechanism. Curr. Opin. Immunol. 31, 51–57 (2014).
David, T., Ling, S. F. & Barton, A. Genetics of immune-mediated inflammatory diseases. Clin. Exp. Immunol. 193, 3–12 (2018).
Olerup, O., Smith, C. I., Bjorkander, J. & Hammarstrom, L. Shared HLA class II-associated genetic susceptibility and resistance, related to the HLA-DQB1 gene, in IgA deficiency and common variable immunodeficiency. Proc. Natl Acad. Sci. USA 89, 10653–10657 (1992).
Maggadottir, S. M. et al. Rare variants at 16p11.2 are associated with common variable immunodeficiency. J. Allergy Clin. Immunol. 135, 1569–1577 (2015).
Fiorillo, E. et al. Autoimmune-associated PTPN22 R620W variation reduces phosphorylation of lymphoid phosphatase on an inhibitory tyrosine residue. J. Biol. Chem. 285, 26506–26518 (2010).
Chew, G. Y. et al. Autoimmunity in primary antibody deficiency is associated with protein tyrosine phosphatase nonreceptor type 22 (PTPN22). J. Allergy Clin. Immunol. 131, 1130–1135.1135.e1131 (2013).
Kalina, T. et al. CD maps-dynamic profiling of CD1-CD100 surface expression on human leukocyte and lymphocyte subsets. Front. Immunol. 10, 2434 (2019).
van Zelm, M. C. et al. Functional antibody responses following allogeneic stem cell transplantation for TP53 mutant pre-B-ALL in a patient with X-linked agammaglobulinemia. Front. Immunol. 10, 895 (2019).
Gaspar, H. B., Lester, T., Levinsky, R. J. & Kinnon, C. Bruton’s tyrosine kinase expression and activity in X-linked agammaglobulinaemia (XLA): the use of protein analysis as a diagnostic indicator of XLA. Clin. Exp. Immunol. 111, 334–338 (1998).
Abolhassani, H. et al. Combined immunodeficiency and Epstein-Barr virus-induced B cell malignancy in humans with inherited CD70 deficiency. J. Exp. Med. 214, 91–106 (2017).
Caorsi, R. et al. CD70 deficiency due to a novel mutation in a patient with severe chronic EBV infection presenting as a periodic fever. Front. Immunol. 8, 2015 (2017).
Izawa, K. et al. Inherited CD70 deficiency in humans reveals a critical role for the CD70-CD27 pathway in immunity to Epstein-Barr virus infection. J. Exp. Med. 214, 73–89 (2017).
Lopez-Herrera, G. et al. Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am. J. Hum. Genet. 90, 986–1001 (2012).
Okeke, E. B. et al. Deficiency of phosphatidylinositol 3-kinase delta signaling leads to diminished numbers of regulatory T cells and increased neutrophil activity resulting in mortality due to endotoxic shock. J. Immunol. 199, 1086–1095 (2017).
Edwards, E. S. J., et al. Predominantly antibody-deficient patients with non-infectious complications have reduced naive B, Treg, Th17 and Tfh17 cells. Front. Immunol. (2019).
Wherry, E. J. et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007).
Xu, W. & Larbi, A. Markers of T cell senescence in humans. Int. J. Mol. Sci. 18, 1742 (2017).
Bercovici, N., Duffour, M. T., Agrawal, S., Salcedo, M. & Abastado, J. P. New methods for assessing T-cell responses. Clin. Diagn. Lab. Immunol. 7, 859–864 (2000).
Ghosh, S. et al. Extended clinical and immunological phenotype and transplant outcome in CD27 and CD70 deficiency. Blood (2020). [Epub ahead of print].
Whittle, J. R. et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88, 4047–4057 (2014).
Wheatley, A. K., Kristensen, A. B., Lay, W. N. & Kent, S. J. HIV-dependent depletion of influenza-specific memory B cells impacts B cell responsiveness to seasonal influenza immunisation. Sci. Rep. 6, 26478 (2016).
Hartley, G. E., et al. Influenza-specific IgG1+ memory B cell numbers increase upon booster vaccination in healthy adults but not in patients with Predominantly Antibody Deficiency. Under review (2020).
Fischer, M. B. et al. A defect in the early phase of T-cell receptor-mediated T-cell activation in patients with common variable immunodeficiency. Blood 84, 4234–4241 (1994).
Li, F. Y. et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature 475, 471–476 (2011).
Chaigne-Delalande, B. et al. Mg2+ regulates cytotoxic functions of NK and CD8 T cells in chronic EBV infection through NKG2D. Science 341, 186–191 (2013).
Tario, J. D. Jr., Conway, A. N., Muirhead, K. A. & Wallace, P. K. Monitoring cell proliferation by dye dilution: considerations for probe selection. Methods Mol. Biol. 1678, 249–299 (2018).
French, M. A. & Harrison, G. Serum IgG subclass concentrations in healthy adults: a study using monoclonal antisera. Clin. Exp. Immunol. 56, 473–475 (1984).
Berneman, A., Belec, L., Fischetti, V. A. & Bouvet, J. P. The specificity patterns of human immunoglobulin G antibodies in serum differ from those in autologous secretions. Infect. Immun. 66, 4163–4168 (1998).
Klinken, E. M. et al. Diversity of XMEN disease: description of 2 novel variants and analysis of the lymphocyte phenotype. J. Clin. Immunol. 40, 299–309 (2020).
Sallusto, F. & Lanzavecchia, A. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur. J. Immunol. 39, 2076–2082 (2009).
Coulter, T. I. & Cant, A. J. The treatment of activated PI3Kdelta syndrome. Front Immunol. 9, 2043 (2018).
Maccari, M. E. et al. Disease evolution and response to rapamycin in activated phosphoinositide 3-kinase delta syndrome: the European Society for Immunodeficiencies-Activated Phosphoinositide 3-Kinase delta Syndrome Registry. Front. Immunol. 9, 543 (2018).
Rao, V. K. et al. Effective “activated PI3Kdelta syndrome”-targeted therapy with the PI3Kdelta inhibitor leniolisib. Blood 130, 2307–2316 (2017).
Cahn, A. et al. Safety, pharmacokinetics and dose-response characteristics of GSK2269557, an inhaled PI3Kdelta inhibitor under development for the treatment of COPD. Pulm. Pharm. Ther. 46, 69–77 (2017).
Kuehn, H. S. et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science 345, 1623–1627 (2014).
Lo, B. et al. Autoimmune disease. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science 349, 436–440 (2015).
Schubert, D. et al. Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat. Med. 20, 1410–1416 (2014).
Alangari, A. et al. LPS-responsive beige-like anchor (LRBA) gene mutation in a family with inflammatory bowel disease and combined immunodeficiency. J. Allergy Clin. Immunol. 130, 481–488 e482 (2012).
Burns, S. O. et al. LRBA gene deletion in a patient presenting with autoimmunity without hypogammaglobulinemia. J. Allergy Clin. Immunol. 130, 1428–1432 (2012).
Revel-Vilk, S. et al. Autoimmune lymphoproliferative syndrome-like disease in patients with LRBA mutation. Clin. Immunol. 159, 84–92 (2015).
Lee, S. et al. Abatacept alleviates severe autoimmune symptoms in a patient carrying a de novo variant in CTLA-4. J. Allergy Clin. Immunol. 137, 327–330 (2016).
van Leeuwen, E. M., Cuadrado, E., Gerrits, A. M., Witteveen, E. & de Bree, G. J. Treatment of Intracerebral Lesions with Abatacept in a CTLA4-Haploinsufficient Patient. J. Clin. Immunol. 38, 464–467 (2018).
Liu, L. et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J. Exp. Med. 208, 1635–1648 (2011).
van de Veerdonk, F. L. et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N. Engl. J. Med. 365, 54–61 (2011).
Boisson-Dupuis, S. et al. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr. Opin. Immunol. 24, 364–378 (2012).
Toubiana, J. et al. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood 127, 3154–3164 (2016).
van Zelm, M. C. et al. Impaired STAT3-dependent upregulation of IL2Ralpha in B cells of a patient with a STAT1 gain-of-function mutation. Front. Immunol. 10, 768 (2019).
Kobbe, R. et al. Common variable immunodeficiency, impaired neurological development and reduced numbers of T regulatory cells in a 10-year-old boy with a STAT1 gain-of-function mutation. Gene 586, 234–238 (2016).
Ramana, C. V., Chatterjee-Kishore, M., Nguyen, H. & Stark, G. R. Complex roles of Stat1 in regulating gene expression. Oncogene 19, 2619–2627 (2000).
Forbes, L. R. et al. Jakinibs for the treatment of immune dysregulation in patients with gain-of-function signal transducer and activator of transcription 1 (STAT1) or STAT3 mutations. J. Allergy Clin. Immunol. 142, 1665–1669 (2018).
Higgins, E. et al. Use of ruxolitinib to successfully treat chronic mucocutaneous candidiasis caused by gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation. J. Allergy Clin. Immunol. 135, 551–553 (2015).
Meesilpavikkai, K. et al. Baricitinib treatment in a patient with a gain-of-function mutation in signal transducer and activator of transcription 1 (STAT1). J. Allergy Clin. Immunol. 142, 328–330 e322 (2018).
Rizzi, M. et al. Outcome of allogeneic stem cell transplantation in adults with common variable immunodeficiency. J. Allergy Clin. Immunol. 128, 1371–1374 e1372 (2011).
Wehr, C. et al. Multicenter experience in hematopoietic stem cell transplantation for serious complications of common variable immunodeficiency. J. Allergy Clin. Immunol. 135, 988–997.e986 (2015).
Fox, T. A. et al. Successful outcome following allogeneic hematopoietic stem cell transplantation in adults with primary immunodeficiency. Blood 131, 917–931 (2018).
Olkinuora, H. et al. T cell regeneration in pediatric allogeneic stem cell transplantation. Bone Marrow Transpl. 39, 149–156 (2007).
Lum, L. G. The kinetics of immune reconstitution after human marrow transplantation. Blood 69, 369–380 (1987).
Naik, S. et al. Adoptive immunotherapy for primary immunodeficiency disorders with virus-specific T lymphocytes. J. Allergy Clin. Immunol. 137, 1498–1505 e1491 (2016).
Castagnoli, R., Delmonte, O. M., Calzoni, E. & Notarangelo, L. D. Hematopoietic stem cell transplantation in primary immunodeficiency diseases: current status and future perspectives. Front. Pediatr. 7, 295 (2019).
Marsh, R. A. et al. Practice pattern changes and improvements in hematopoietic cell transplantation for primary immunodeficiencies. J. Allergy Clin. Immunol. 142, 2004–2007 (2018).
Gennery, A. R. et al. Transplantation of hematopoietic stem cells and long-term survival for primary immunodeficiencies in Europe: entering a new century, do we do better? J. Allergy Clin. Immunol. 126, 602–610.e601-611 (2010).
Bezu, L., Chuang, A. W., Liu, P., Kroemer, G. & Kepp, O. Immunological effects of epigenetic modifiers. Cancers 11 (2019).
Campos-Sanchez, E. & Martinez-Cano, J. Del Pino Molina, L., Lopez-Granados, E. & Cobaleda, C. Epigenetic deregulation in human primary immunodeficiencies. Trends Immunol. 40, 49–65 (2019).
Del Pino-Molina, L. et al. Impaired CpG demethylation in common variable immunodeficiency associates with B cell phenotype and proliferation rate. Front. Immunol. 10, 878 (2019).
Titus, A. J., Gallimore, R. M., Salas, L. A. & Christensen, B. C. Cell-type deconvolution from DNA methylation: a review of recent applications. Hum. Mol. Genet. 26, R216–R224 (2017).
Park, J. et al. Interferon signature in the blood in inflammatory common variable immune deficiency. PLoS ONE 8, e74893 (2013).
Rodenburg, R. J. The functional genomics laboratory: functional validation of genetic variants. J. Inherit. Metab. Dis. 41, 297–307 (2018).
Stavnezer-Nordgren, J., Kekish, O. & Zegers, B. J. Molecular defects in a human immunoglobulin kappa chain deficiency. Science 230, 458–461 (1985).
Bottaro, A., Cariota, U. & DeMarchi, M. & Carbonara, A.O. Pulsed-field electrophoresis screening for immunoglobulin heavy-chain constant-region (IGHC) multigene deletions and duplications. Am. J. Hum. Genet. 48, 745–756 (1991).
Ferrari, S. et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc. Natl Acad. Sci. USA 98, 12614–12619 (2001).
Zhang, K. H., Marsh, A. & Jordan, R. M.B. Identification of a phosphoinositide 3-kinase (PI-3K) p110 delta (PIK3CD) deficient individual. J. Clin. Immunol. 33, 673–674 (2013).
van Montfrans, J. M. et al. CD27 deficiency is associated with combined immunodeficiency and persistent symptomatic EBV viremia. J. Allergy Clin. Immunol. 129, 787–793.e786 (2012).
Salzer, E. et al. B-cell deficiency and severe autoimmunity caused by deficiency of protein kinase C delta. Blood 121, 3112–3116 (2013).
Salzer, E. et al. Early-onset inflammatory bowel disease and common variable immunodeficiency-like disease caused by IL-21 deficiency. J. Allergy Clin. Immunol. 133, 1651–1659.e1612 (2014).
Kotlarz, D. et al. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J. Exp. Med. 210, 433–443 (2013).
Wiseman, D. H. et al. A novel syndrome of congenital sideroblastic anemia, B-cell immunodeficiency, periodic fevers, and developmental delay (SIFD). Blood 122, 112–123 (2013).
Zhou, Q. et al. A hypermorphic missense mutation in PLCG2, encoding phospholipase Cgamma2, causes a dominantly inherited autoinflammatory disease with immunodeficiency. Am. J. Hum. Genet. 91, 713–720 (2012).
Acknowledgements
The authors are indebted to A/Prof Robert Stirling, A/Prof Paul Cameron, Dr. Josh Chatelier, Ms. Pei M. Aui and Ms. Fiona Hore-Lacy for clinical, laboratory and administrative support, and to E/Prof Jennifer Rolland for critical reading of the manuscript. This work is supported by The Jeffrey Modell Foundation and the Australian National Health and Medical Research Council (NHMRC; Senior Research Fellowship 1117687 to M.C.v.Z.).
Author information
Authors and Affiliations
Contributions
E.S.J.E. and M.C.v.Z. conceptualized and wrote the manuscript. J.J.B., S.O., and R.E.O.H. provided critical feedback and commented on manuscript drafts. All authors approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Edwards, E.S.J., Bosco, J.J., Ojaimi, S. et al. Beyond monogenetic rare variants: tackling the low rate of genetic diagnoses in predominantly antibody deficiency. Cell Mol Immunol 18, 588–603 (2021). https://doi.org/10.1038/s41423-020-00520-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-020-00520-8