Abstract
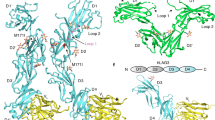
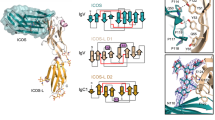
Leukocyte immunoglobulin (Ig)-like receptors (LILRs), also known as CD85 and immunoglobulin-like transcripts (ILTs), play pivotal roles in regulating immune responses. These receptors define an immune checkpoint that immune therapy can target. Through cis or trans interactions with human leukocyte antigen (HLA)-G, the two most abundantly expressed inhibitory LILRs, LILRB1, and LILRB2 (LILRB1/2, also known as CD85j/d and ILT2/4), are involved in immunotolerance in pregnancy and transplantation, autoimmune diseases, and immune evasion by tumors. Although the discrete domains of LILRB1/2 are clear, the assembly mode of the four extracellular Ig-like domains (D1, D2, D3, and D4) remains unknown. Previous data indicate that D1D2 is responsible for binding to HLA class I (HLA-I), but the roles of D3D4 are still unclear. Here, we determined the crystal structure of the four Ig-like domain LILRB2 and four-domain LILRB1 in complex with HLA-G1. The angles between adjacent domains and the staggered assembly of the four domains suggest limited flexibility and limited plasticity of the receptors during ligand binding. The complex structure of four-domain LILRB1 and HLA-G1 supports the model that D1D2 is responsible for HLA-I binding, while D3D4 acts as a scaffold. Accordingly, cis and trans binding models for HLA-I binding to LILRB1/2 are proposed. The geometries of LILRB1/2 in complex with dimeric and monomeric HLA-G1 suggest the accessibility of the dimeric receptor, which in turn, transduces more inhibitory signals. The assembly of LILRB1/2 and its binding to HLA-G1 could aid in the design of immune regulators and benefit immune interference.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The accession numbers for the atomic coordinates and diffraction data reported in this paper are PDB 6AED (crystal structure of LILRB2) and 6AEE (crystal structure of LILRB1/HLA-G1 complex), respectively.
References
Brown, D., Trowsdale, J. & Allen, R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 64, 215–225 (2004).
Takeda, K. & Nakamura, A. Regulation of immune and neural function via leukocyte Ig-like receptors. J. Biochem. 162, 73–80 (2017).
Zhang, J. et al. Leukocyte immunoglobulin-like receptors in human diseases: an overview of their distribution, function, and potential application for immunotherapies. J. Leukoc. Biol. 102, 351–360 (2017).
Colonna, M. et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 186, 1809–1818 (1997).
Shiroishi, M. et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl Acad. Sci. USA 100, 8856–8861 (2003).
Merlo, A. et al. Inhibitory receptors CD85j, LAIR-1, and CD152 down-regulate immunoglobulin and cytokine production by human B lymphocytes. Clin. Diagn. Lab. Immunol. 12, 705–712 (2005).
Favier, B., LeMaoult, J., Lesport, E. & Carosella, E. D. ILT2/HLA-G interaction impairs NK-cell functions through the inhibition of the late but not the early events of the NK-cell activating synapse. FASEB J. 24, 689–699 (2010).
Ponte, M. et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc. Natl Acad. Sci. USA. 96, 5674–5679 (1999).
Masuda, A., Nakamura, A., Maeda, T., Sakamoto, Y. & Takai, T. Cis binding between inhibitory receptors and MHC class I can regulate mast cell activation. J. Exp. Med. 204, 907–920 (2007).
Mori, Y. et al. Inhibitory immunoglobulin-like receptors LILRB and PIR-B negatively regulate osteoclast development. J. Immunol. 181, 4742–4751 (2008).
Roberti, M. P. et al. Overexpression of CD85j in TNBC patients inhibits Cetuximab-mediated NK-cell ADCC but can be restored with CD85j functional blockade. Eur. J. Immunol. 45, 1560–1569 (2015).
Barkal, A. A. et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. 19, 76–84 Nat. Immunol. (2017).
Kang, X. et al. Inhibitory leukocyte immunoglobulin-like receptors: Immune checkpoint proteins and tumor sustaining factors. Cell Cycle. 15, 25–40 (2016).
Willcox, B. E., Thomas, L. M. & Bjorkman, P. J. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat. Immunol. 4, 913–919 (2003).
Shiroishi, M. et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc. Natl Acad. Sci. USA. 103, 16412–16417 (2006).
Yang, Z. & Bjorkman, P. J. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc. Natl Acad. Sci. USA. 105, 10095–10100 (2008).
Dulberger, C. L. et al. Human leukocyte antigen F presents peptides and regulates immunity through interactions with NK cell receptors. Immunity. 46, 1018–1029 e1017 (2017).
Mohammed, F. et al. The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status. Oncotarget. 8, 54160–54172 (2017).
Chapman, T. L., Heikeman, A. P. & Bjorkman, P. J. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 11, 603–613 (1999).
Huang, J. et al. HLA-B*35-Px-mediated acceleration of HIV-1 infection by increased inhibitory immunoregulatory impulses. J. Exp. Med. 206, 2959–2966 (2009).
Gao, X. et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344, 1668–1675 (2001).
Steinle, A. et al. Motif of HLA-B*3503 peptide ligands. Immunogenetics. 43, 105–107 (1996).
Lichterfeld, M. et al. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 204, 2813–2824 (2007).
Yang, Y., Huang, J., Toth, I., Lichterfeld, M. & Yu, X. G. Mutational escape in HIV-1 CTL epitopes leads to increased binding to inhibitory myelomonocytic MHC class I receptors. PLoS ONE. 5, e15084 (2010).
Nam, G. et al. Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding. Protein Cell. 4, 761–770 (2013).
Lichterfeld, M. & Yu, X. G. The emerging role of leukocyte immunoglobulin-like receptors (LILRs) in HIV-1 infection. J. Leukoc. Biol. 91, 27–33 (2012).
Chapman, T. L., Heikema, A. P., West, A. P. & Bjorkman, P. J. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2). Immunity. 13, 727–736 (2000).
Willcox, B. E. et al. Crystal structure of LIR-2 (ILT4) at 1.8 Å: differences from LIR-1 (ILT2) in regions implicated in the binding of the human cytomegalovirus class I MHC homolog UL18. BMC Struct. Biol. 2, 6 (2002).
Shiroishi, M. et al. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J. Biol. Chem. 281, 10439–10447 (2006).
Colonna, M. et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 186, 1809–1818 (1997).
Chen, Y. et al. Crystal structure of myeloid cell activating receptor leukocyte Ig-like receptor A2 (LILRA2/ILT1/LIR-7) domain swapped dimer: molecular basis for its non-binding to MHC complexes. J. Mol. Biol. 386, 841–853 (2009).
Cheng, H. et al. Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance. J. Biol. Chem. 286, 18013–18025 (2011).
Zhang, W. et al. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell. 1, 459–467 (2010).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Acknowledgements
We thank the staff of BL17U beamline at SSRF. We are grateful to Zheng Fan from the Institute of Microbiology Chinese Academy of Sciences (CAS) for technical assistance with the SPR experiments. This work was supported by the Strategic Priority Research Program of CAS (grant no. XDA12020358), the National Basic Research Program (973) of China (grant no. 2015CB910503), and the National Natural Science Foundation of China (NSFC, grant nos 31502078 and 31390432). Q.W. is supported by the Youth Innovation Promotion Association CAS (grant no. 2018119). H.S. is supported by the Young Elite Scientist Sponsorship Program by CAST (2016QNRC001) and the Youth Innovation Promotion Association CAS (2017117). G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (Grant no. 81621091).
Author contributions
Y.S. and G.F.G. initiated and coordinated the project. Q.W., H.S., Y.S., J.Y. and G.F.G. designed the experiments. Q.W. and H.S. conducted the experiments with the assistance of G.N. Q.W. obtained diffractable complex crystals of LILRB1 and HLA-G1, and H.S. obtained diffractable crystals of free LILRB2. J.Q. solved the crystal structures. Q.W., H.S., H.C., Y.S., J.Y. and G.F.G. analyzed the data. Q.W. wrote the manuscript. H.S., S.T., J.W., M.F., Z.T., X.C., Z.A., J.Y. and G.F.G revised the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Q., Song, H., Cheng, H. et al. Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex. Cell Mol Immunol 17, 966–975 (2020). https://doi.org/10.1038/s41423-019-0258-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-019-0258-5
Keywords
This article is cited by
-
Harnessing the potential of HLA-G in cancer therapy: advances, challenges, and prospects
Journal of Translational Medicine (2024)
-
Natural LILRB1 D1-D2 variants show frequency differences in populations and bind to HLA class I with various avidities
Immunogenetics (2022)
-
Structural basis for RIFIN-mediated activation of LILRB1 in malaria
Nature (2020)