Abstract
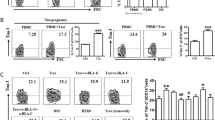
Macrophages are crucial for a successful pregnancy, and malfunctions of decidual macrophages correlate with adverse pregnancy outcomes, such as spontaneous abortion and preeclampsia. Previously, decidual macrophages were often thought to be a single population. In the present study, we identified three decidual macrophage subsets, CCR2−CD11cLO (CD11clow, ~80%), CCR2−CD11cHI (CD11chigh, ~5%), and CCR2+CD11cHI (CD11chigh, 10–15%), during the first trimester of human pregnancy by flow cytometry analysis. CCR2−CD11cLO macrophages are widely distributed in the decidua, while CCR2−CD11cHI and CCR2+CD11cHI macrophages are primarily detected close to extravillous trophoblast cells according to immunofluorescence staining. According to RNA sequencing bioinformatics analysis and in vitro functional studies, these three subsets of macrophages have different phagocytic capacities. CCR2+CD11cHI macrophages have pro-inflammatory characteristics, while the CCR2−CD11cHI population is suggested to be anti-oxidative and anti-inflammatory due to its high expression of critical heme metabolism-related genes, suggesting that these two subsets of macrophages maintain an inflammatory balance at the leading edge of trophoblast invasion to facilitate the clearance of pathogen infection as well as maintain the homeostasis of the maternal-fetal interface. The present study physiologically identifies three decidual macrophage subsets. Further clarification of the functions of these subsets will improve our understanding of maternal-fetal crosstalk in the maintenance of a healthy pregnancy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Erlebacher, A. Immunology of the maternal-fetal interface. Annu. Rev. Immunol. 31, 387–411 (2013).
Wallace, A. E., Whitley, G. S., Thilaganathan, B. & Cartwright, J. E. Decidual natural killer cell receptor expression is altered in pregnancies with impaired vascular remodeling and a higher risk of pre-eclampsia. J. Leukoc. Biol. 97, 79–86 (2015).
Fu, B. et al. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity 47, 1100–1113 (2017). e1106.
Zhang, J. et al. Human dNK cell function is differentially regulated by extrinsic cellular engagement and intrinsic activating receptors in first and second trimester pregnancy. Cell. Mol. Immunol. 14, 203–213 (2017).
Aluvihare, V. R., Kallikourdis, M. & Betz, A. G. Regulatory T cells mediate maternal tolerance to the fetus. Nat. Immunol. 5, 266–271 (2004).
Lash, G. E. et al. Decidual macrophages: key regulators of vascular remodeling in human pregnancy. J. Leukoc. Biol. 100, 315–325 (2016).
Abrahams, V. M., Kim, Y. M., Straszewski, S. L., Romero, R. & Mor, G. Macrophages and apoptotic cell clearance during pregnancy. Am. J. Reprod. Immunol. 51, 275–282 (2004).
Egashira, M. et al. F4/80+ macrophages contribute to clearance of senescent cells in the mouse postpartum uterus. Endocrinology 158, 2344–2353 (2017).
Hamilton, S. et al. Macrophages infiltrate the human and rat decidua during term and preterm labor: evidence that decidual inflammation precedes labor. Biol. Reprod. 86, 39 (2012).
Wang, H. et al. Role of decidual CD14(+) macrophages in the homeostasis of maternal-fetal interface and the differentiation capacity of the cells during pregnancy and parturition. Placenta 38, 76–83 (2016).
Care, A. S. et al. Macrophages regulate corpus luteum development during embryo implantation in mice. J. Clin. Invest. 123, 3472–3487 (2013).
Murray, P. J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Gustafsson, C. et al. Gene expression profiling of human decidual macrophages: evidence for immunosuppressive phenotype. PLoS ONE 3, e2078 (2008).
Jaiswal, M. K. et al. V-ATPase upregulation during early pregnancy: a possible link to establishment of an inflammatory response during preimplantation period of pregnancy. Reproduction 143, 713–725 (2012).
Zhang, Y. H., He, M., Wang, Y. & Liao, A. H. Modulators of the balance between M1 and M2 macrophages during pregnancy. Front. Immunol. 8, 120 (2017).
Ning, F., Liu, H. & Lash, G. E. The role of decidual macrophages during normal and pathological pregnancy. Am. J. Reprod. Immunol. 75, 298–309 (2016).
Heyward, C. Y. et al. The decidua of preeclamptic-like BPH/5 mice exhibits an exaggerated inflammatory response during early pregnancy. J. Reprod. Immunol. 120, 27–33 (2017).
Tsao, F. Y., Wu, M. Y., Chang, Y. L., Wu, C. T., Ho, H. N. M1 macrophages decrease in the deciduae from normal pregnancies but not from spontaneous abortions or unexplained recurrent spontaneous abortions. J. Formos. Med. Assoc. (2017).
Xu, Y. et al. An M1-like macrophage polarization in decidual tissue during spontaneous preterm labor that is attenuated by rosiglitazone treatment. J. Immunol. 196, 2476–2491 (2016).
Gomez-Lopez, N., StLouis, D., Lehr, M. A., Sanchez-Rodriguez, E. N. & Arenas-Hernandez, M. Immune cells in term and preterm labor. Cell. Mol. Immunol. 11, 571–581 (2014).
Fu, B. et al. CD11b and CD27 reflect distinct population and functional specialization in human natural killer cells. Immunology 133, 350–359 (2011).
Zeng, W. et al. Distinct transcriptional and alternative splicing signatures of decidual CD4+ T cells in early human pregnancy. Front. Immunol. 8, 682 (2017).
Piccinni, M. P. et al. Defective production of both leukemia inhibitory factor and type 2 T-helper cytokines by decidual T cells in unexplained recurrent abortions. Nat. Med. 4, 1020–1024 (1998).
Houser, B. L., Tilburgs, T., Hill, J., Nicotra, M. L. & Strominger, J. L. Two unique human decidual macrophage populations. J. Immunol. 186, 2633–2642 (2011).
Tilburgs, T. et al. Human HLA-G+ extravillous trophoblasts: Immune-activating cells that interact with decidual leukocytes. Proc. Natl Acad. Sci. USA 112, 7219–7224 (2015).
Li, Y. H. et al. The Galectin-9/Tim-3 pathway is involved in the regulation of NK cell function at the maternal-fetal interface in early pregnancy. Cell. Mol. Immunol. 13, 73–81 (2016).
Hung, J. H. & Weng, Z. Analyzing MicroarrayData. Cold Spring Harb. Protoc. 2017, pdbprot093112 (2017).
Law, C. W., Alhamdoosh, M., Su, S., Smyth, G. K. & Ritchie, M. E. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res 5, 1408 (2016).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Rhee, S. Y., Wood, V., Dolinski, K. & Draghici, S. Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 9, 509–515 (2008).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Pirooznia, M., Nagarajan, V. & Deng, Y. GeneVenn - a web application for comparing gene lists using Venn diagrams. Bioinformation 1, 420–422 (2007).
Wang, Z. et al. Interferon-gamma inhibits nonopsonized phagocytosis of macrophages via an mTORC1-c/EBPbeta pathway. J. Innate Immun. 7, 165–176 (2015).
Kurihara, T., Warr, G., Loy, J. & Bravo, R. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J. Exp. Med. 186, 1757–1762 (1997).
Zheng, Y. et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 540, 458–461 (2016).
Zhang, Y. et al. Human trophoblast cells induced MDSCs from peripheral blood CD14(+) myelomonocytic cells via elevated levels of CCL2. Cell. Mol. Immunol. 13, 615–627 (2016).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Elomaa, O. et al. Structure of the human macrophage MARCO receptor and characterization of its bacteria-binding region. J. Biol. Chem. 273, 4530–4538 (1998).
Arredouani, M. et al. The scavenger receptor MARCO is required for lung defense against pneumococcal pneumonia and inhaled particles. J. Exp. Med. 200, 267–272 (2004).
Thelen, T. et al. The class A scavenger receptor, macrophage receptor with collagenous structure, is the major phagocytic receptor for Clostridium sordellii expressed by human decidual macrophages. J. Immunol. 185, 4328–4335 (2010).
Aldape, M. J., Bryant, A. E. & Stevens, D. L. Clostridium sordellii infection: epidemiology, clinical findings, and current perspectives on diagnosis and treatment. Clin. Infect. Dis. 43, 1436–1446 (2006).
Callewaert, L. & Michiels, C. W. Lysozymes in the animal kingdom. J. Biosci. 35, 127–160 (2010).
Gozzelino, R., Jeney, V. & Soares, M. P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 50, 323–354 (2010).
Naito, Y., Takagi, T. & Higashimura, Y. Heme oxygenase-1 and anti-inflammatory M2 macrophages. Arch. Biochem. Biophys. 564, 83–88 (2014).
Gibbings, S. L. et al. Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol. 57, 66–76 (2017).
Hess, A. P. et al. Decidual stromal cell response to paracrine signals from the trophoblast: amplification of immune and angiogenic modulators. Biol. Reprod. 76, 102–117 (2007).
Ozen, M., Zhao, H., Lewis, D. B., Wong, R. J. & Stevenson, D. K. Heme oxygenase and the immune system in normal and pathological pregnancies. Front. Pharmacol. 6, 84 (2015).
Acknowledgements
We thank Xili Zhu and Shiwen Li for confocal image capture, Hua Qin for FACS, Can Peng and Lei Sun for TEM, and Tingting Wu for the phagocytosis experiment. We thank Meijing Wang, Rong Jing, and Jinglei Zhai for collecting samples from the hospital, and we thank Yong Zhao, Jingpian Peng, Yanling Wang, and Jay. C. Cross for discussion. This study was supported by grants from the National Natural Science Foundation of China (81490741 and 81401224) and the Ministry of Science and Technology of the People’s Republic of China (2016YFC1000208 and 2017YFC1001401).
Authors’ contributions
X.J. performed the experiments. H.W. and X.J. designed the study and interpreted the data. M.L. collected samples from the hospital. X.J. wrote the original manuscript. M.R.D. and H.W. modified the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jiang, X., Du, MR., Li, M. et al. Three macrophage subsets are identified in the uterus during early human pregnancy. Cell Mol Immunol 15, 1027–1037 (2018). https://doi.org/10.1038/s41423-018-0008-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-018-0008-0
Keywords
This article is cited by
-
Decidual macrophage: a reversible role in immunotolerance between mother and fetus during pregnancy
Archives of Gynecology and Obstetrics (2024)
-
Estrogen-sensitive activation of SGK1 induces M2 macrophages with anti-inflammatory properties and a Th2 response at the maternal–fetal interface
Reproductive Biology and Endocrinology (2023)
-
Investigation into the role of the MITA-TRIM38 interaction in regulating pyroptosis and maintaining immune tolerance at the maternal-fetal interface
Cell Death & Disease (2023)
-
Infections at the maternal–fetal interface: an overview of pathogenesis and defence
Nature Reviews Microbiology (2022)
-
Tim-3+ decidual Mφs induced Th2 and Treg bias in decidual CD4+T cells and promoted pregnancy maintenance via CD132
Cell Death & Disease (2022)