Summary
TSPAN family of proteins are generally considered to assemble as multimeric complexes on the plasma membrane. Our previous work uncovered that TSPAN8 can translocate into the nucleus as a membrane-free form, a process that requires TSPAN8 palmitoylation and association with cholesterol to promote its extraction from the plasma membrane and subsequent binding with 14-3-3θ and importin-β. However, what upstream signal(s) regulate(s) the nuclear translocation of TSPAN8, the potential function of TSPAN8 in the nucleus, and the underlying molecular mechanisms all remain unclear. Here, we demonstrate that, epidermal growth factor receptor (EGFR) signaling induces TSPAN8 nuclear translocation by activating the kinase AKT, which in turn directly phosphorylates TSPAN8 at Ser129, an event essential for its binding with 14-3-3θ and importin ß1. In the nucleus, phosphorylated TSPAN8 interacts with STAT3 to enhance its chromatin occupancy and therefore regulates transcription of downstream cancer-promoting genes, such as MYC, BCL2, MMP9, etc. The EGFR–AKT–TSPAN8–STAT3 axis was found to be hyperactivated in multiple human cancers, and associated with aggressive phenotype and dismal prognosis. We further developed a humanized monoclonal antibody hT8Ab4 that specifically recognizes the large extracellular loop of TSPAN8 (TSPAN8-LEL), thus being able to block the extraction of TSPAN8 from the plasma membrane and consequently its nuclear localization. Importantly, both in vitro and in vivo studies demonstrated an antitumor effect of hT8Ab4. Collectively, we discovered an unconventional function of TSPAN8 and dissected the underlying molecular mechanisms, which not only showcase a new layer of biological complexity of traditional membrane proteins, but also shed light on TSPAN8 as a novel therapeutic target for refractory cancers.
Similar content being viewed by others
Introduction
Tetraspanins are cell-surface glycoproteins that are characterized by four hydrophobic membrane-spanning domains. Thirty-four structurally highly conserved tetraspanin members have been identified in mammals, including clusters of differentiation (CD)-related protein 9 (CD9), CD37, CD53, CD81, CD82, CD151, and tetraspanin-8 (TSPAN8, encoded by TSPAN8 gene).1,2,3 Previous studies suggest that tetraspanins may aggregate with one another, and dynamically recruit a multitude of transmembrane or signal-transducing molecules at the cell surface or membranous organelle, forming specialized multimeric complexes referred as “tetraspanin web” or “tetraspanin-enriched microdomains” (TEM).4 In these complexes, tetraspanins act as molecular scaffolds to modulate the properties of the recruited proteins, including integrins, immunoglobulin superfamily members, adhesion molecules, ectoenzymes (ectopeptidases and metalloproteases) and intracellular signaling molecules such as receptor-tyrosine kinases (RTKs).5,6,7,8 Depending on various associated molecules, tetraspanins are involved in numerous biological processes including immune responses, cell adhesion, motility, proliferation, membrane dynamics and fusion, as well as viral infection and cancer metastasis.9,10,11,12,13,14
Recently, we and others showed that TSPAN8 play important roles in tumor progression in a variety of human tumors.11,15,16,17,18,19,20,21 Liu et al. reported that elevated TSPAN8 expression on serum extracellular vesicles is associated with distant metastasis of non-small cell lung cancer.22 Of note, we found that TSPAN8 expression is upregulated in breast cancer stem cells.23 Mechanistically, TSPAN8 directly interacts with PTCH1 and inhibits the proteasome-mediated degradation of the SHH/PTCH1 complex through recruitment of ATXN3 deubiquitinating enzyme, contributing to hedgehog signaling (Hh) activation and therapeutic resistance.23 This is consistent with a previous report that Lgr5+TSPAN8hi subset represents a deeply quiescent mammary stem cell population, with transcriptome strikingly similar to that of claudin-low tumors.24 Wang et al. demonstrated that TSPAN8 is a biomarker of migrating pancreatic cancer-initiating cells.25 In addition, we recently identified sex determining region Y-related HMG-box (SOX) as a critical transcriptional regulator of TSPAN8 expression in response to EGF stimulation.26 These findings demonstrate important roles of TSPAN8 signaling cascade during cancer progression and metastasis, highlighting TSPAN8 as a key target for cancer therapy.
By serendipity, we previously found that TSPAN8 is also present in the cytoplasm and inside the nucleus.27 The nuclear localization of TSPAN8 can be detected in multiple cancer cells, indicating a general phenomenon with pathological implications of this protein in the nucleus. Specifically, we found that the palmitoylation of TSPAN8 and subsequently binding with cholesterol are important for its structural protection in a full-length form and extraction from the membrane. As a result, the TSPAN8–cholesterol complex binds to 14-3-3θ and importin β to achieve nuclear translocation. However, the underlying molecular mechanism of TSPAN8 nuclear transport, especially its upstream inducing signal(s), and its functional role in the nucleus remain unclear.
Here, we report that EGF–EGFR signaling orchestrates TSPAN8 nuclear translocation. Mechanistically, EGF–EGFR signaling activates AKT, which in turn phosphorylates TSPAN8 at Ser129; pTSPAN8S129 binds to the molecular chaperone 14-3-3θ and is translocated into the nucleus, where it interacts with the transcription factor STAT3 to promote the transcription of downstream target genes during cancer progression and metastasis. After showing a strong clinical relevance of nuclear TSPAN8 with refractory cancers, we further generated a humanized monoclonal antibody for TSPAN8, which inhibits tumor progression by blocking the extraction of TSPAN8 from the plasma membrane and therefore its nuclear localization. Together, our study, in combination with previous reports,27,28 establishes a nuclear role for plasma membrane proteins such as TSPAN8, and exemplifies a targeting strategy for therapeutic purpose against such unconventional pathology.
Results
Clinical relevance and pathological association of nuclear TSPAN8 with tumorigenesis
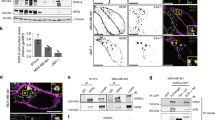
Previously, we determined the subcellular localization of TSAPN8 and found that TSPAN8 could be detected in nucleus in multiple cell lines.27 To evaluate the biological consequence and importance of TSPAN8 nuclear translocation, we set out to investigate the pathological significance of nuclear TSPAN8 expression. To this end, we employed a commercial anti-TSPAN8 antibody with verified specificity both in vitro and in vivo (Supplementary information, Fig. S1), and performed immunohistochemical staining (IHC) analyses in clinical specimens of primary human breast cancer (BrCa, n = 95) (Fig. 1a; Supplementary information, Fig. S2a), gastric cancer (GC, n = 60) (Supplementary information, Fig. S2b), colorectal cancer (CRC, n = 60) (Supplementary information, Fig. S2c), and pancreatic ductal adenocarcinoma (PDAC, n = 60) (Supplementary information, Fig. S2d). Our results showed that nuclear TSPAN8 expression was significantly upregulated in a variety of tumor tissues compared to their paired normal tissues (Fig. 1b; Supplementary information, Fig. S2e). Nuclear TSPAN8 levels were positively correlated with aggressive features of advanced stage of cancer (Fig. 1c), triple-negative subtype (TNBC) (Fig. 1d), increased tumor size (Fig. 1e), lymph node involvement (Fig. 1f), distant metastasis (Fig. 1g), and mesenchymal phenotype (Fig. 1h) in BrCa. Moreover, we noticed an obvious increase in the ratio of cells with apparent nuclear staining of TSPAN8 (TSPAN8Nuc) to cells with TSPAN8 predominately stained in the cytoplasm (TSPAN8Cyto) in BrCa tissues when compared to the paired normal tissues (Fig. 1i). Then, we used Image-Pro Plus Version 6.2 software to separate the BrCas into a high nuclear TSPAN8 expression group (The integrated optical density (IOD) > median) and a low TSPAN8 nuclear expression group (IOD ≤ median). Of note, BrCa patients with high TSPAN8 nuclear expression had significantly shorter overall survival (hazard ratio (HR) = 0.4803, 95% confidence interval (CI) = 0.3101 to 0.7439, Fig. 1j). In addition, multivariate analysis indicated nuclear TSPAN8 as an independent prognostic factor for BrCa patients (Supplementary information, Table. S1). Taken together, these results confirmed the nuclear localization of TSPAN8 and further revealed a pathological association of nuclear TSPAN8 with tumorigenesis.
a Representative IHC images (magnification ×2, ×10, and ×20) of the TSPAN8 staining (from two patients) in a microarray consisting of n = 95 BrCa patients. BrCa, breast cancer; Bar, 100 μm. b Nuclear localization of TSPAN8 is significantly increased in BrCa. Quantification of total TSPAN8 and nuclear TSPAN8 from BrCa IHC microarray is shown. Staining (%) is calculated using Image J by comparing total and nuclear TSPAN8 IHC integrated optical density (IOD) with total IOD values extracted within the image. Paired t-test. c Nuclear TSPAN8 level (%) is positively correlated with TNM stages in BrCa patients. One-way ANOVA analysis with Bonferroni post hoc test. d Nuclear TSPAN8 level (%) is specifically increased in TNBC subtype. Quantification of nuclear TSPAN8 in BrCa patients with different molecular subtypes (luminal A/B, Her2+, TNBC) is shown. One-way ANOVA analysis with Bonferroni post hoc test. TNBC, triple-negative breast cancer. e–g Nuclear TSPAN8 level (%) is positively correlated with tumor aggressive status. Quantification of nuclear TSPAN8 in BrCa patients with different tumor size (T, e), lymph node involvement (N, f) or distant metastasis (M, g) is shown. Student’s t-test. h Nuclear TSPAN8 level (%) is positively correlated with mesenchymal phenotype. Quantification of nuclear TSPAN8 in BrCa patients with E-cad-positive (Ecad+) and E-cad-negative (Ecad−) expression is shown. Student’s t-test. i Ratio of cells with nuclear TSPAN8 staining to cells with cytosol TSPAN8 staining. Paired t-test. j High level of nuclear TSPAN8 predicates dismal prognosis in BrCa. Kaplan-Meier survival analysis of patients with high or low levels of TAPAN8 is shown. Log-rank test.
EGF–EGFR signaling orchestrates TSPAN8 nuclear translocation
Given the above findings of nuclear TSPAN8 localization and its potential role in tumorigenesis, we then interrogated the molecular mechanism through which TSPAN8 was translocated from the plasma membrane to the nucleus. To this end, we first probed the upstream signal(s) that induce the nuclear translocation of TSPAN8. In view of the hyperactivation of EGF–EGFR signaling commonly observed in malignant tumors with more aggressive features,29,30,31,32,33,34 we speculated that EGF–EGFR signaling may modulate TSPAN8 function in a subcellular-compartment-dependent manner. To test this hypothesis, we performed immunofluorescence analysis of nuclear TSPAN8 expression in BrCa cells with or without EGF treatment. To our surprise, the results showed a marked increase of nuclear accumulation of TSPAN8 in both MCF-7 (Fig. 2a) and MDA-MB-231 (Fig. 2b) human BrCa cells upon EGF treatment. Consistent with the increased nuclear expression of TSPAN8 in TNBC subtype BrCa tissues (Fig. 1d), we found that the percentage of cells with apparent nuclear staining of TSPAN8 was higher for MDA-MB-231 cell line as compared with MCF-7 line (Fig. 2a, b). Considering that the physiological concentration of EGF ranges from 10 to 100 ng/mL,35 we further determined the dose-dependent effect of EGF towards promoting the nuclear translocation of TSPAN8. Indeed, the result of immunofluorescence analysis revealed that EGF treatment dose-dependently enhanced the nuclear translocation of TSPAN8 (Fig. 2c). Moreover, we observed that the protein levels of nuclear TSPAN8 were significantly increased 15 min post EGF treatment (100 ng/mL), peaking at 60 min and then descending toward the basal level at 120 min (Fig. 2d). Next, to determine the relative abundance of nuclear TSPAN8 after EGF treatment, we performed cellular compartment fractionation assay to isolate cell membrane, cytoplasm, and nuclear fractions to quantitatively analyze the relative abundance of TSPAN8 in each compartment of MDA-MB-231 cells via western blotting. Though nuclear TSPAN8 only accounted for a small part (9%) of total TSPAN8 protein at basal level, the ratio of this part was dramatically increased to nearly 22% upon EGF treatment (Fig. 2e). Interestingly, this analysis revealed that the ratio of cytoplasmic TSPAN8 was decreased from 61% to 50%, whereas the ratio of plasma membrane-associated TSPAN8 was almost unchanged (Fig. 2e), indicating that EGF promotes TSPAN8 cytoplasm-to-nucleus translocation.
a, b EGF treatment promotes TSPAN8 nuclear translocation. MCF-7 (a) or MDA-MB-231 cells (b) were treated with or without EGF for 60 min and TSPAN8 subcellular localization was examined via IFA assay. Bar, 10 μm. Quantification was from three independent experiments. Student’s t-test. c EGF promotes TSPAN8 nuclear translocation in a dose-dependent manner. MCF-7 cells were treated with indicated dose of EGF and subjected to IFA analysis. d Time-dependent analysis of nuclear TSPAN8 by western blotting after EGF treatment in MCF-7 cells. e EGF mainly drives TSPAN8 cytoplasm-to-nucleus translocation. MDA-MB-231 cells were untreated or treated with EGF for 60 min before being processed for cellular compartment fractionation assay. Left panel, representative blotting images of indicated proteins. Right panel, Ratio of protein in each compartment was quantified from three independent experiments. Student’s t-test. f Schematic presentation of using Mem-TurboID to label and track the mobilization of membranous TSPAN8 to nucleus in response to EGF in MDA-MB-231 cells. g EGF-induced nuclear TSPAN8 is derived from plasma membrane. Following Mem-TurboID labeling, western blotting was performed to detect relative abundance of biotinylated TSPAN8 in membrane and nucleus compartments at different time points after treatment with 100 ng/mL EGF. h EGF treatment does not affect TSPAN8 membrane extraction. MDA-MB-231 cells were treated with different doses of EGF and processed as in g.
Our previous finding suggest that site-specific palmitoylation of TSPAN8 is required for its binding with cholesterol and subsequent membrane extraction.27 Consistently, we found that EGF treatment did not change the palmitoylation status of TSPAN8 (Supplementary information, Fig. S3a). Considering that TSPAN8 undergoes constitutive palmitoylation in the cells, we next attempted to determine whether the nuclear TSPAN8 induced by EGF is derived originally from plasma membrane or simply from newly synthesized protein in cytoplasm. To this end, we employed plasma membrane restrained, proximality-dependent labeling strategy (Mem-TurboID) (Supplementary information, Fig. S3b)36,37 to pre-label the membranous TSPAN8 by biotinylation before EGF treatment. After removal of biotin, we treated the cells with 100 ng/mL EGF for another one hour, and collected both cell membrane and nuclear fractions at indicated time points for streptavidin immunoprecipitation and examined the presence of biotinylated TSPAN8 in each compartment by western blotting (Fig. 2f). Interestingly, we found that the level of biotinylated TSPAN8 protein in membrane fraction was decreased, whereas nuclear biotinylated TSPAN8 was increased in a time-dependent manner after EGF treatment (Fig. 2g), suggesting that nuclear TSPAN8 was originally derived from cell membrane. Next, to further determine whether EGF may also affect TSPAN8 membrane extraction, we treated MDA-MB-231 cells with different dose of EGF after initial biotin labeling and collected samples at a fixed time points (60 min) to examine the residual membrane biotinylated TSPAN8. The rational is that if EGF affect TSPAN8 membrane extraction, the residual membrane biotinylated TSPAN8 should be changed by EGF treatment in a dose-dependent manner. Interestingly, we reproducibly observed a dose-dependent nuclear accumulation of biotinylated TSPAN8, however, this analysis failed to detect any reduction of membrane-associated biotinylated TSPAN8 at the same time point (Fig. 2h), which was consistent with our observation from cellular compartment assay (Fig. 2e). Taken together, these results demonstrated that EGF treatment did not alter TSPAN8 membrane extraction, but promoted cytoplasm-to-nucleus translocation of extracted membranous TSPAN8.
The kinase AKT directly phosphorylates TSPAN8 at Ser129 upon EGF–EGFR signaling
Given that the EGF–EGFR signaling triggers activation of diverse cytoplasmic tyrosine kinases,38 we speculated that tyrosine kinases may also play a vital role in mediating TSPAN8 nuclear translocation. To test this hypothesis, we treated MDA-MB-231 cells with commercial tyrosine kinase inhibitors (TKIs) such as Afatinib, Erlotinib or AEF788 before EGF treatment, and then examined TSPNA8 nuclear localization frequency via IFA assay. We found that treatment with these TKIs efficiently blocked EGF-induced nuclear translocation of TSPAN8 (Fig. 3a; Supplementary information, Fig. S3c). As TSPAN8 can exist in the cytoplasm as both membrane-associated (P100) and membrane-free (S100) forms, we then examined the effects of TKIs on distribution of TSPAN8 in different subcellular compartments upon EGF treatment. Interestingly, we found that TKIs treatment only resulted in enhanced accumulation of TSPAN8 in S100 fraction, without affecting TSPAN8 localization in cell membrane and P100 fractions (Supplementary information, Fig. S3d), suggesting that EGF mainly promotes TSPAN8 cytosol-to-nucleus translocation.
a Treatment of TKIs inhibit EGF-induced TSPAN8 nuclear translocation. MDA-MB-231 cells pre-treated with indicated inhibitors were further treated with EGF for 60 min before processing for TSPAN8 staining. b Identification of AKT as a potential TSPAN8-interacting protein via BioID assay. c EGF treatment promotes TSPAN8–AKT interaction. MDA-MB-231 cells were treated with EGF for indicated time points followed by TSPAN8 Co-IP assay. d AKT interacts with TSPAN8 at cytoplasm. MDA-MB-231 cells treated with EGF for 60 min were subjected to cytoplasmic fraction-based AKT immunoprecipitation. e AKT phosphorylates TSPAN8 in vitro. Purified AKT kinase was incubated with recombinant GST or GST-TSPAN8 for 30 min before being subjected to western blotting analysis via indicated antibodies. f MS-based phospho-proteomics analysis to reveal key residues of TSPAN8 phosphorylated by AKT. g AKT phosphorylates TSPAN8 at Ser129 in vitro. Purified AKT kinase was incubated with indicated recombinant GST proteins for 30 min before subjecting to western blotting analysis. h Sequence alignment of TSPAN8 Ser129 between indicated species. i AKT inhibition attenuates EGF-induced TSPAN8 phosphorylation at Ser129. MDA-MB-231 cells were transiently transfected with indicated constructs for 30 h. After that, cells were pre-treated with or without AKT inhibitor MK-2206 for 2 h followed by EGF treatment for another 60 min. Cells were then subjected to Flag immunoprecipitation assay and analyzed via western blotting using indicated antibodies.
We then applied proximity labeling (BioID)-based mass spectrometry (MS) assay,39 which can efficiently label the transient protein–protein interaction with the addition of biotin substrate, to characterize the TSPAN8-binding proteins (Supplementary information, Table 2). Interestingly, this analysis revealed enhanced interactions of TSPAN8 with AKT upon EGF treatment compared with control (Fig. 3b). Subsequently, we validated the MS results by performing TSPAN8 co-immunoprecipitation (Co-IP) assay at different time points after EGF treatment. Results clearly showed that the association between TSPAN8 and AKT was strongly increased following EGF treatment (Fig. 3c). As AKT has also been reported to translocate into the nucleus,40,41 we then performed reciprocal AKT Co-IP assay in both cytoplasm and nuclear fractions to identify where the interaction occurs. We detected a robust AKT–TSPNA8 interaction in the cytoplasm which was further enhanced upon EGF treatment (Fig. 3d). However, we failed to detect such interaction in nucleus (Supplementary information, Fig. S3e).
Since AKT is a kinase downstream of the EGF–EGFR signaling, we then asked whether AKT directly phosphorylates TSPAN8. To test this possibility, we then carried out in vitro kinase assay with an anti-pan phospho-Tyr/Ser antibody. As shown in Fig. 3e, active AKT was indeed able to phosphorylate wild-type TSPAN8 in vitro, indicating TSPAN8 as a bona-fide substrate for AKT. Next, we performed MS analysis to identify the specific sites of TSPAN8 phosphorylated by AKT upon EGF treatment. MS analysis indicated S129, T131 and T196 of TSPAN8 are possible sites that are phosphorylated upon EGF treatment (Fig. 3f). Then, we used a point-mutation strategy to verify these candidate sites of TSPAN8. It is noteworthy that the TSPAN8 S129A mutation, but not T131A and T196A mutations, abolished the phosphorylation of TSPAN8 by AKT in vitro (Fig. 3g). Sequence alignment of TSPAN8 Ser129 revealed that this residue is conserved from dog to human (Fig. 3h). To further examine whether this is the case in vivo, we treated MDA-MB-231 cells expressing TSPAN8 (wildtype (WT) and mutants) with EGF. Consistently, we observed elevated TSPAN8 phosphorylation in WT-, T131A-, and S196A-expressing cells, which was greatly attenuated by further incubation with AKT inhibitor MK2206 (Fig. 3i). Nevertheless, the S129A mutation almost blocked EGF-induced TSPAN8 phosphorylation regardless of MK2206 treatment (Fig. 3i). Taken together, these results demonstrated that the kinase AKT directly interacts with and phosphorylates TSPAN8 at Ser129 upon EGF–EGFR signaling.
Phosphorylated TSPAN8 enters the nucleus via enhanced binding to 14-3-3θ and importin ß1
To investigate the potential role of Ser129 phosphorylation of TSPAN8 (hereafter referred as pTSPAN8S129) during EGF–EGFR signaling-induced nuclear translocation of TSPAN8, we generated two mutants: a TSPAN8S129A variant that can no longer be phosphorylated by AKT; a TSPAN8S129D variant that mimics its constitutive phosphorylation by AKT under EGF/EGFR signaling activation. We overexpressed WT and mutant TSPAN8 in MCF-7 cells, and examined the nuclear and cytoplasmic fractions of various forms of TSPAN8 by immunofluorescence assay (IFA). If TSPAN8 phosphorylation at S129 is essential for its nuclear translocation, we would expect decreased nuclear fraction of the TSPAN8S129A mutant but increased nuclear fraction of the TSPAN8S129D mutant when compared to that of WT TSAPN8 (hereafter referred as TSAPN8WT). Indeed, results showed that the TSPAN8S129A mutant was barely detectable in the nucleus; while the TSPAN8S129D mutant was distributed throughout the cell including the nucleus (Fig. 4a).
a pTSPAN8S129 enhances the nuclear localization of TSPAN8. MCF-7 cells were transfected with indicated plasmids for 30 h before processing for IFA assay. Bar, 10 μm. Quantification was performed for data from three experiments. Student’s t-test. b TSPAN8 phosphorylation promotes breast cancer cell proliferation. MCF-7 cells transfected with indicated plasmids were measured by CellTiter Luminescent-based assay at indicated time points. c EGF promotes breast cancer cell colony formation via pTSPAN8S129. MDA-MB-231 cells expressing TSPAN8WT, TSPAN8S129A or TSPAN8S129D constructs were subjected to colony formation assay in the absence or presence of EGF treatment. Right panel, colonies were counted 10 d after treatment. Student’s t-test. d levels of pTSPAN8S129 in cells after EGF treatment for different time. e, f pTSPAN8S129 level is upregulated in BrCa tissues. Representative image shows the staining of pTSPAN8S129 on BrCa tissue microarrays (e). Bar, 100 μm. Quantification of pTSPAN8S129 level for 95 BrCa patients is also shown (f). Staining (%) is calculated using Image J by comparing pTSPAN8S129 IHC IOD with total IOD obtained within the image. Paired t-test. g, h pTSPAN8S129 positively correlates with tumor aggressive phenotype. Quantification of pTSPAN8S129 in BrCa patients with different TNM stages (g) or molecular subtypes (h) is shown. Staining (%) is calculated as in f. One-way ANOVA analysis with Bonferroni post hoc test.
It is noteworthy that, our previous report indicate that TSPAN8 does not translocate into nuclei in the form of vesicles, but in a form of TSPAN8–cholesterol complex.27 In this case, the hydrophobic transmembrane domain can be protected by cholesterol and the Ser129 site of TSPAN8 can be phosphorylated by AKT. To validate this hypothesis, we further examined the cytosol-localized pTSPAN8 level immediately after EGF stimulation (30 min), and found that S100-enriched pTSPAN8S129 was dramatically increased upon EGF treatment (Supplementary information, Fig. S4a). To further show that disruption of pTSPAN8S129 mainly affects the membrane-free form of TSPAN8, we also compared the relative abundance of Flag-tagged TSPAN8WT and TSPAN8S129A in S100 and P100 fractions after EGF treatment. Western blotting analysis revealed much more TSPAN8S129A in the S100 fraction as compared with TSAPN8WT (Supplementary information, Fig. S4b), which phenocopies the effect of TKIs on S100-loalized TSPAN8 (Supplementary information, Fig. S3d). These results suggest that EGF–AKT–pTSPAN8S129 axis mainly mediates cytoplasmic-to-nuclear translocation of membrane-fee TSPAN8.
It is widely accepted that 14-3-3 proteins play a pivotal role in protein subcellular localization and nuclear import. Our previous work demonstrated that R18, a 14-3-3 peptide inhibitor, blocks the translocation of TSPAN8 into nuclei, and knockdown of 14-3-3θ, but not 14-3-3ζ, blocks nuclear translocation of TSPAN8.27 Consistent with the observation of EGF-induced TSPAN8 nuclear translocation, we found EGF treatment can dose-dependently promote interactions between TSPAN8 with 14-3-3θ and importin ß1 (Supplementary information, Fig. S4c). We then asked whether TSPAN8 phosphorylation might affect its association with 14-3-3θ and thus be involved in TSPAN8 nuclear translocation. Indeed, reciprocal Co-IP assay showed that TSPAN8S129D displayed much stronger, whereas TSPAN8S129A showed greatly impaired binding ability to 14-3-3θ in MDA-MB-231 cells (Supplementary information, Fig. S4d).
Consistent with our previous findings, we next used the siRNA silencing approach to specifically knock down 14-3-3θ and importin ß1 in MDA-MB-231 cells and then re-analyzed the TSPAN8 expression in nuclear and cytoplasmic fractions by both immunofluorescence and immunoblotting assays. Results showed that depletion of either 14-3-3θ or importin ß1 specifically inhibited EGF-promoted TSPAN8 accumulation in the nuclear fraction (Supplementary information, Fig. S4e, f). Taken together, these data indicate that AKT-mediated phosphorylation at Ser129 promotes TSPAN8 nuclear translocation, and does so by facilitating its binding with 14-3-3θ and importin ß1.
pTSPAN8S129 causes more aggressive cancer phenotypes than TSPAN8WT
We then assessed the pathological importance of pTSPAN8S129 in tumor progression. To this end, we first measured the cell proliferation abilities of MCF-7 cells transfected with TSPAN8WT, TSPAN8S129A or TSPAN8S129D. Intriguingly, we found that cells overexpressing the TSPAN8S129D mutant showed significantly increased proliferation rate than cells overexpressing TSPAN8WT or the TSPAN8S129A mutant (Fig. 4b), displaying a strong positive correlation with the nuclear localization pattern of TSPAN8 (Fig. 4a). Consistent with the growth-promoting effect of pTSPAN8S129, we found cells overexpressing TSPAN8S129D mutant showed higher colony formation efficiency (Supplementary information, Fig. S5a) and invasion ability when compared to cells overexpressing TSPAN8WT (Supplementary information, Fig. S5b). Next, to further validate the pro-proliferative functions of pTSPAN8S129, we treated MDA-MB-231 cells expressing indicated TSPAN8 constructs with or without EGF, and evaluated their colony formation abilities. Consistently, we observed that MDA-MB-231 cells overexpressing TSPAN8S129D mutant displayed higher colony formation efficiency than those expressing TSPAN8WT and TSPAN8S129A. Notably, further EGF treatment dramatically promoted colony formation of TSPAN8WT-expressing, but not TSPAN8S129A-expressing cells (Fig. 4c).
To further characterize the pathological function of TSPAN8 phosphorylation by AKT, we utilized a polyclonal antibody specifically recognizing pTSPAN8S129. By using this antibody, we found that the levels of endogenous pTSPAN8S129 in MDA-MB-231 cells was progressively increased and peaking at 60 min after EGF treatment (Fig. 4d). In addition, we found that in S100 fraction Ser129 phosphorylation of TSPAN8 was dramatically increased 30 min after EGF treatment (Supplementary information, Fig. S4a). We then used this antibody to examine pTSPAN8S129 in BrCa tissues previously used for TSPAN8 analysis. As expected, the levels of pTSPAN8S129 were significantly increased in BrCa tissues compared with the paired normal tissues (Fig. 4e, f). Furthermore, pTSPAN8S129 was predominantly localized in the nucleus and was positively correlated with TSPAN8 nuclear expression (Supplementary information, Fig. S5c). Moreover, the elevated levels of pTSPAN8S129 were clearly associated with mesenchymal feature (Supplementary information, Fig. S5d), advanced stage (Fig. 4g), and TNBC subtype (Fig. 4h). Of note, high levels of pTSPAN8S129 was correlated with shorter overall survival time of BrCa patients (P = 0.0003, Supplementary information, Fig. S5e). Taken together, these results demonstrated a positive correlation between pTSPAN8S129 and its nuclear localization as well as the important pathological role for pTSPAN8S129 during tumor progression.
Phosphorylated TSPAN8 interacts with the transcription factor STAT3 in the nucleus
To further gain insights into the mechanism underlying the function of TSPAN8 in the nuclues, we analyzed our data of TSPAN8 BioID assay and noted that more peptides of transcription factor STAT3 were identified in our TSPAN8-BioID assay after EGF treatment (Figs. 3b, 5a). Subsequent Co-IP assays further substantiated the EGF-enhanced interaction between TSPAN8 and STAT3 (Fig. 5b). Moreover, in vitro pull-down assay using purified proteins of Flag-TSPAN8 and Myc-STAT3 identified the direct interaction between TSPAN8 and STAT3 (Fig. 5c; Supplementary information, Fig. S6a). Next, we investigated whether TSPAN8–STAT3 interaction is mediated by pTSPAN8S129 upon EGF treatment. Indeed, we reproducibly detected the EGF-enhanced TSPAN8–STAT3 interaction in MDA-MB-231 cells, however, such interaction was greatly attenuated for TSPAN8S129A mutant regardless of EGF stimulation (Fig. 5d). On the contrary, TSPAN8S129D mutant displayed much enhanced STAT3-binding capacity compared with TSPAN8WT even in the absence of EGF (Fig. 5d), suggesting that pTSPAN8S129 and the nuclear import of TSPAN8 mainly mediate TSPAN8 interaction with STAT3. In addition, considering that both 14-3-3θ and STAT3 can bind to TSPAN8 upon EGF stimulation, we next examined whether these two factors cooperatively or competitively interact with TSPAN8 in the nucleus. Surprisingly, we found that ectopic expression of STAT3 to enhance its interaction with TSPAN8 resulted in gradually decreased association between 14-3-3θ and TSPAN8 (Fig. 5e), implying a competitive fashion of STAT3 and 14-3-3θ to interact with TSPAN8. Collectively, these results indicate that upon phosphorylation at S129, TSPAN8 is transferred into nucleus where it directly interacts with STAT3.
a Coverage of STAT3 peptides in control or EGF-stimulated cells by TSPAN8-BioID assay. b EGF promotes STAT3–TSPN8 interaction. Co-IP analysis of endogenous STAT3 with TSPAN8 in MDA-MB-231 cells treated with or without EGF. c STAT3 directly interacts with TSPAN8 in vitro. Purified proteins from eukaryote cell free system were subjected to Flag pulldown assay and analyzed via western blotting. d EGF promotes STAT3–TSPAN8 interaction via pTSPAN8S129. MDA-MB-231 cells transfected with indicated Flag-tagged constructs were subjected to Co-IP assay with or without EGF treatment. The interaction between TSPAN8 and endogenous STAT3 was detected. e Ectopic STAT3 expression attenuates TSPAN8–importin interaction. MDA-MB-231 cells were co-transfected with Flag-TSPAN8 and Myc-STAT3 with different doses for 36 h before processing for nuclear Co-IP assay. f, g Nuclear STAT3 level is upregulated in BrCa tissues. Representative image shows the staining of STAT3 on BrCa tissue microarrays (f). Bar, 100 μm. Quantification of STAT3 from BrCa IHC microarray is also shown (g). The percentage of staining (%) is calculated using Image J by comparing nuclear STAT3 with total STAT3 IHC IOD. Paired t-test. h Nuclear TSPAN8 and STAT3 double positive predicates worst prognosis in BrCa. Survival analysis of patients with TSPAN8/STAT3 at high or low levels is shown. i, j Nuclear STAT3 positively correlates with nuclear TSPAN8 and pTSPAN8S129 levels in BrCa tissues. Chi-square test of the correlation between the levels of STAT3 and TSPAN8/pTSPAN8 in the nucleus using data form IHC microarray is shown.
To determine the clinical relevance of the TSPAN8–STAT3 interaction, we next performed IHC staining to examine pTSPAN8S129, and the nuclear expression of TSPAN8 and STAT3 in serial sections of human primary BrCa specimens (n = 95) using antibodies with validated specificities. Our results revealed that the nuclear expression of STAT3 in the BrCa tissues was higher than that in the paired normal tissues (Fig. 5f, g). It is worth noting that BrCa patients with high protein levels of both TSPAN8 and STAT3 in the nucleus show worse prognosis compared to those with high nuclear expression of either TSPAN8 or STAT3 (Fig. 5h). Further Chi-square analysis demonstrated a positive correlation between the expression levels of nuclear TSPAN8 and nuclear STAT3 (Fig. 5i), as well as one between pTSPAN8S129 and nuclear STAT3 (Fig. 5j) in BrCa specimens. Taken together, these results indicate that pTSPAN8S129 interacts with STAT3 in the nucleus to promote cancer progression.
pTSPAN8S129 stabilizes STAT3 on target gene loci genome-wide to regulate transcription
Given our finding of the TSPAN8–STAT3 interaction, we hypothesized that nuclear TSPAN8 may form a complex with STAT3 on DNA to regulate target gene transcription. To test this possibility, we first performed RNA-seq analysis for TSPAN8-depletion MCF-7 cells upon EGF treatment. After alignment to the human reference genome and normalization, we identified a total of 20,312, 21,536 and 20,611 transcripts for control, EGF and siTSPAN8 + EGF cells, respectively. This information was used to evaluate transcriptional differences between the WT and TSPAN8-depleted cells after EGF treatment. Gene set variation analysis (GSVA) clearly revealed a significantly decreased enrichment of JAK_STAT3 signaling upon TSPAN8 depletion (Fig. 6a). 622 uniquely expressed transcripts of STAT3_Gene_Set (1366)42 were identified in TSPAN8 + EGF groups (Fig. 6b). In addition, we performed RNA-seq transcriptome analysis for MCF-7 cells overexpressing TSPAN8S129D; and Gene set enrichment analysis (GSEA) revealed a significant enrichment of STAT3 target genes when compared with cells expressing TSPAN8WT (Fig. 6c). These results indicate that nuclear TSPAN8 indeed can regulate STAT3 activity and target gene transcription.
a RNA-Seq analysis revealed potential signaling pathways regulated by TSPAN8. MCF-7 cells were transfected with control or TSPAN8 siRNAs and were further treated with or without EGF before processed for RNA-Seq and GSVA analysis. b Venn diagram with differentially transcribed mRNAs in siTSPAN8 + EGF and STAT3 Gene Set. Numbers of individual mRNA transcripts, EGF vs siTSPAN8 + EGF (blue), and STAT3 Gene Set (grey) are indicated. c Ectopic expression of TSPAN8S129D activates STAT3 signaling. GSEA showing significant enrichment of STAT3 target genes in TSPAN8S129D-overexpressing group. d Heat map representing TSPAN8 and STAT3 binding sites located on promoters and enhancers upon EGF treatment. e Venn diagram of peaks identified by anti-TSPAN8 and anti-STAT3 antibodies. f Identification of STAT3 motif in TSPAN8 binding sequences. g Absolute distance of TSPAN8 peaks, STAT3 peaks, or overlapping TSPAN8/STAT3 peaks to the nearest TSS. h Linear correlation between the signals of TSPAN8 and STAT3 peaks in the 2163 shared binding sites. r, the coefficient of determination of the two correlations. i EGF promotes TSPAN8 binding to target gene promoters. MDA-MB-231 cells were treated with EGF for indicated time points before being subjected to ChIP-qPCR assay using primers spanning the human MYC, BCL2 or MMP9 locus. One-way ANOVA analysis with Bonferroni post hoc test. j STAT3 inhibition blocks TSPAN8 binding to MYC, BCL2 or MMP9 promoters. MDA-MB-231 cells were treated with EGF in combination with BAY2353 or WP1066 before being subjected to ChIP-qPCR assay. One-way ANOVA analysis with Bonferroni post hoc test.
Subsequently, we assessed at genome-wide the association between TSPAN8 and STAT3 by performing chromatin immunoprecipitation followed by sequencing (ChIP-seq) assays with anti-TSPAN8 and anti-STAT3 antibodies (Fig. 6d). Intriguingly, a total of 2136 peaks at genome-wide level were identified by both antibodies, accounting for 42% (2136/5212) of the peaks identified with anti-TSPAN8 antibody (Fig. 6e). Intriguingly, motif analysis revealed that overlapped peaks mainly locate at SOX10, TCF3, SMAD4, and STAT3/4/5 motifs (Supplementary information, Fig. S6b), and that the canonical STAT3-binding motif ranks No. 1 within TSPAN8-related binding sequences (Fig. 6f). Moreover, most of these shared binding sites are located on active enhancers, indicating a genome-wide association between TSPAN8 and STAT3. Analysis of the distribution of TSPAN8- or STAT3-binding sites relative to genes annotated in the human genome revealed that only a very small fraction of peaks were mapped close (< 1 kb) to transcription start sites (TSSs), whereas most peaks were located farther than 10 kb from the closest TSS (Fig. 6g). Notably, the signals of TSPAN8 peaks were found to be positively correlated with those of STAT3 peaks (Fig. 6h).
To corroborate our ChIP-seq analyses, we next examined the occupancy of TSPAN8 and STAT3 on specific target gene loci by ChIP-qPCR assay. First, we found that EGF treatment can promote both TSPAN8 (Fig. 6i) and STAT3 binding (Supplementary information, Fig. S6c) to promotes of MYC, BCL2 and MMP9 in a time-dependent manner. Consistent with the positive correlation between pTSPAN8S129 and the TSPAN8–STAT3 interaction in nucleus (Fig. 5d), the TSPAN8S129D mutant exhibited much enhanced, whereas TSPAN8S129A showed reduced such binding ability to BCL2 promoter compared with TSPAN8WT (Supplementary information, Fig. S6d). Besides, ectopic expression of TSPAN8S129D led to much stronger enrichment of STAT3 on BCL2 promoter than TSPAN8WT, which could further be increased by EGF stimulation (Supplementary information, Fig. S6e), indicating that TSPAN8 positively regulates the chromatin association of STAT3.
Accordingly, we also examined the essential role of STAT3 in the binding of TSPAN8 to chromatin. In this regard, though EGF can time-dependently promote TSPAN8 association with chromatin, such binding ability was dramatically reduced in STAT3-depleted MDA-MB-231 cells (Supplementary information, Fig. S6f). Indeed, co-treatment with commercial STAT3 inhibitor WP106643 or BAY235244 blocked the EGF-induced TSPAN8 binding to promotes of MYC, BCL2 and MMP9 (Fig. 6j), indicating that STAT3 plays an essential role in recruiting TSPNA8 to chromatin in nucleus. Taken together, these results demonstrated that nuclear TSPAN8 forms a complex with STAT3 to coregulate their occupancy on target gene loci and therefore target gene transcription.
pTSPAN8S129 promote tumor progression via STAT3-dependent gene expression
To further assess the functional importance of the TSPAN8–STAT3 axis in breast tumorigenesis, we verified the oncogenic role of pTSPAN8S129 in xenograft tumor model. To this end, around 1 × 106 MCF-7 cells were first injected into the breast fat pad of Balb/c nude mice to incubate tumor formation. After that, the mice were intratumorally injected with lentivirus particles engineered with TSPAN8WT, TSPAN8S129A or TSPAN8S129D (Supplementary information, Fig. S6g). Compared to control vector group, we found that mice receiving TSPAN8WT particles developed tumors rapidly, while the tumors in mice injected with TSPAN8S129D particles grew even larger (Fig. 7a, b). In contrast, tumors formed in mice receiving TSPAN8S129A particles were smaller than those in mice injected with TSPAN8WT (Fig. 7a, b). These observations were further confirmed by IHC analysis showing a much higher protein level of the cell proliferation marker Ki67 in tumor tissues of TSPAN8S129D group but a relatively lower level of Ki67 for the TSPAN8S129A group when compared to the TSPAN8WT group (Fig. 7c). Consistently, we observed by RNA scope assay that the mRNA level of BCL2 was significantly increased in the TSPAN8S129D group tumor tissues but decreased in the TSPAN8S129A group tumors when compared to the TSPAN8WT group (Fig. 7d). In addition, we found that depletion of TSPAN8 in MCF-7 cells dramatically reduced its tumor formation ability (Fig. 7e), Ki67-positive cell rate (Fig. 7f), as well as BCL2 expression level (Fig. 7g) in tumor tissues compared with WT group. Importantly, further reconstitution using TSPAN8WT only partially, whereas TSPAN8S129D more profoundly rescued the tumor formation abilities and expression of proliferation markers (Fig. 7e–g). Collectively, these results indicate a key role of pTSPAN8S129 and nuclear localization in promoting cancer progression.
a, b Nuclear TSPAN8 promotes tumor growth in vivo. Xenograft tumor growth of MCF-7 cells after infection with the indicated lentivirus (n = 6) for 3 weeks. Representative images were presented (a) and tumor size was quantified (b). One-way ANOVA analysis with Bonferroni post hoc test. c Ki67 staining of tumors shown in panel a. Bar, 10 μm. Quantification of Ki67-positive cells from three independent images was performed and the mean value was listed below images. d Nuclear TSPAN8 promotes BCL2 transcription in vivo. RNA scope-immunohistochemistry of BCL2 in tumors shown in panel a. Bar, 10 μm. e Depletion of TSPAN8 suppresses tumor formation. Xenograft tumor growth of TSPAN8-knockdown MCF-7 cells after infection with the indicated lentivirus (n = 6). Tumor volumes were determined 3 weeks after injection. One-way ANOVA analysis with Bonferroni post hoc test. f, g Depletion of TSPAN8 reduces tumor cell proliferation and BCL2 transcription in vivo. Ki67 staining (f), as well as of RNA scope of BCL2 (g) were performed for tumors shown in panel e. Bar, 10 μm. h, i Nuclear TSPAN8 promotes tumor growth via STAT3. Xenograft tumor growth of TSPAN8S129D-overexpressing MCF-7 cells after transfection with siSTAT3 (n = 6). Tumor images were shown (h) and tumor volumes were quantified (i). Student’s t-test. j STAT3 depletion reduces TSPAN8-mediated BCL2 transcription. RNA scope of BCL2 was performed for tumors shown in panel h. Bar, 10 μm.
To further interrogate the functional dependency of nuclear TSPAN8 on STAT3, we injected mice with MCF-7 cells transfected with TSPAN8S129D in the presence or absence of STAT3 siRNA (Fig. 7h, i). The results showed that tumors in TSPAN8S129D group were substantially decreased in size when STAT3 was depleted (Fig. 7h, i). Further RNA scope analyses also showed that TSPAN8S129D-induced mRNA expression of BCL2 was apparently reduced by STAT3 depletion (Fig. 7j). Together, these findings strongly suggest that nuclear TSPAN8 promotes tumor progression in a STAT3-dependent manner.
A humanized anti-TSPAN8 monoclonal antibody therapy for treating refractory cancers
Based on the mechanistic understanding of the tumor-promoting function of nuclear TSPAN8, we wondered whether blocking the nuclear translocation of TSPAN8 would be therapeutically beneficial. To test this possibility, we applied phage display technology and a synthetic human scFv library to generate a human monoclonal antibody via screen a panel of purified antigens targeting the LEL of TSPAN8 (Fig. 8a; Supplementary information, Fig. S7a). Among the 8 initial candidates, we noted that #4, #5, and #6 displayed relative stronger binding specificity to TSPAN8 via either ELISA (Fig. 8b) or flow cytometry analysis (Fig. 8c), and that #4 showed better staining property in recognizing exogenously GFP-TSPAN8 via IFA assay (Fig. 8d; Supplementary information, Fig. S7b), and thus #4 was termed as hT8Ab4 and was chosen for further characterization.
a Flowchart of phage display to screen inhibitory antibody against TSPAN8. b Phage ELISA assay that detects the binding of TSPAN8 with indicated antibodies. c Flow cytometric analysis of TSPAN8 using the indicated antibodies. d hT8Ab4 displays excellent staining specificity. MDA-MB-231 cells were transfected with GFP-TSPAN8 for 30 h before immunofluorescent staining using the indicated antibodies. e hT8Ab4 treatment limits cancer cell migration. Cell migration assay of MCF-7 cells after treatment with anti-TSPAN8 antibody. One-way ANOVA analysis with Bonferroni post hoc test. f TBNC cells are more sensitive to hT8Ab4 treatment. MCF-10A, MDA-MB-231 and 4175 cells were treated with or without hT8Ab4 and subjected to CellTiter Luminescent-based assay at indicated time points. g TSPAN8high cancer cells are more sensitive to hT8Ab4 treatment. Scatter plots for hT8Ab4 IC50 versus TSPAN8 mRNA levels in a panel of cancer cell lines. h, i hT8Ab4 dose-dependently inhibits tumor growth. Xenograft tumor growth of MDA-MB-231 cells after intratumoral injection with indicated dose of anti-TSPAN8 antibody (n = 4). Representative images on day 15 are shown (h) and tumor volumes on day 0, day 3, day 7, day 14, and day21 (i) were quantified after injection. One-way ANOVA analysis with Bonferroni post hoc test. j hT8Ab4 treatment reduces BCL2 transcription in vivo. RNA scope analysis of BCL2 was performed for tumors shown in panel i. Bar, 10 μm. k hT8Ab4 treatment suppresses TSPAN8 nuclear translocation in vivo. TSPAN8 staining was performed for tumor tissues shown in panel i. Bar, 10 μm. l High dose hT8Ab4 treatment shows no drug toxicity. Healthy nude mice were intraperitoneally injected with 50 mg/kg or 100 mg/kg of hT8Ab4 antibody for three successive days, and body weight were measured at day 1, day 5, day 10, day 15, day 20, and day 25 post-injection (n = 5).
First, surface plasmon resonance (SPR) assay revealed that the hT8Ab4 antibody can dose dependently bind to the purified antigen TSPAN8-LEL-Fc in vitro (Supplementary information, Fig. S7c). Besides, melting point test showed that the antibody displayed well-tolerant protein stability below 70 °C (Supplementary information, Fig. S7d). Moreover, western blotting analysis revealed that hT8Ab4 antibody displayed excellent specificity using either TSPNA8-ectopic expression or depleted samples (Supplementary information, Fig. S7e). Importantly, to specifically show that hT8Ab4 can recognize surface TSPAN8, we incubated MDA-MB-231 cells with different doses of hT8Ab4. Flow cytometry analysis revealed that the mean fluorescence intensity was greatly increased with the increase of antibody concentration (Supplementary information, Fig. S7f), and the calculated binding EC50 was 23 nM (Supplementary information, Fig. S7g). Collectively, these biophysical or biomedical results demonstrated the high-quality of our humanized antibody.
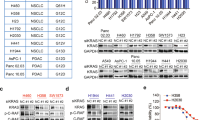
Next, we evaluated whether hT8Ab4 exhibit anti-tumor activity in vitro using different cancer cell lines. First, treatment with hT8Ab4 dramatically attenuated the EGF-stimulated TSPAN8 nuclear translocation in MDA-MB-231 cells (Supplementary information, Fig. S8a). Accordingly, treatment with hT8Ab4 could dose-dependently reduce the breast cancer cell growth (Supplementary information, Fig. S8b, c) and cell invasion (Fig. 8e). Moreover, consistent with the aberrant TSPAN8 nuclear localization in TBNC tumors, we found that MDA-MB-231 cells as well as its derivative 4175 cells were more sensitive to hT8Ab4-mediated anti-proliferation effect than MCF-10A cells (Fig. 8f). To further evaluate the antitumor efficacy of hT8Ab4 towards cells with different expression levels of TSPAN8, we examined the effect of hT8Ab4on growth of 10 cancer cell lines derived from breast, gastric, and colon tumors by measuring the individual IC50 values of hT8Ab4 in these cells. Our results showed that the expression levels of TSPAN8 in the treated cells were generally positively correlated with the sensitivity of these tumor cells towards hT8Ab4 (Fig. 8g).
Then, we testified the efficacy of hT8Ab4 against human breast cancer xenografts in nude mice. Administration of hT8Ab4 via intratumoral injection (1 or 10 mg/kg) significantly inhibited the growth of xenograft tumors derived from MDA-MB-231 cell line. Imaging with Tumor Paint (chlorotoxin: Cy5.5), a tumor-tracking molecular bioconjugate, showed decreased tumor size on day 14 in both 1 or 10 mg/kg hT8Ab4-treated mice (Fig. 8h). Analysis of gross tumor growth after treatment demonstrated a significant reduction in tumor burden in hT8Ab4-treated mice (Fig. 8i). Consistently, IHC analysis and RNA-scope analysis revealed decreased expression of proliferation marker Ki67 (Supplementary information, Fig. S8c) and BCL-2 (Fig. 8j), respectively, in tumor sections treated with hT8Ab4as compared with control tumors. In line with the in vitro data, we observed that nuclear TSPAN8 staining was gradually reduced in tumor tissues derived from hT8Ab4-treated mice (Fig. 8k). Nevertheless, no significant loss of weights or obvious signs of toxicity, such as decreased activity, appetite reduction, or lethargy, were observed during the efficacy study or in a separate toxicity study, in which animals were administrated via intravenous injection with much higher dose of antibodies (50 or 100 mg/kg) during the experimental procedure, as shown by body weight and gross anatomical examination of organs (Fig. 8i).
Finally, to better validate the link between anti-tumor activity of hT8Ab4 with pTSPAN8S129 and STAT3 dependency. We detected the hT8Ab4 sensitivity using TNBC patient-derived organoids (PDOs) generated following our previously published method.45 First, we infected these PDOs with lentiviral particles expressing TSPAN8WT, TSPAN8S129A, and TSPAN8S129D and examined their effect on PDOs formation. Consistent with the tumor-promoting role of TSPAN8, the number of organoids was significantly increased in TSPAN8WT group as compared with mock group, which was further enhanced in TSPAN8S129D group (Supplementary information, Fig. S8d). Moreover, hT8Ab4 treatment markedly suppressed the organoids formation for TSPAN8WT and TSPAN8S129D group, but showed attenuated inhibition effect for TSPAN8S129A group (Supplementary information, Fig. S8d). Further CCK8 assay revealed very similar trends related to hT8Ab4 killing effect towards PDOs expressing indicated proteins (Supplementary information, Fig. S8e). These results validated that the anti-tumor activity of therapeutic hT8Ab4 antibody is at least largely dependent on blocking the nuclear translocation of TSPAN8. In addition, consistent with our notion that STAT3 acts downstream of TSPAN8 to promote tumor growth, we found that STAT3 inhibitor treatment can phenocopy the anti-tumor activity of our monoclonal antibody hT8Ab4, resulting in reduced numbers of organoids (Supplementary information, Fig. S8f) and lower cells viability (Supplementary information, Fig. S8g). Of note, our therapeutic antibody hT8Ab4 displayed comparable inhibitory effect as WP1066, but was better than BAY2353.
Taken together, these results confirm the pathological role of nuclear TSPAN8/pTSPAN8S129–STAT3 in tumorigenesis, and further provide proof-of-concept to therapeutically target nuclear TSPAN8 for cancer treatment.
Discussion
Emerging evidence has shown that some protein receptors originally thought localized only on the plasma membrane can shuttle into the nucleus under certain cellular contexts.46,47,48,49,50,51,52,53 Thus, the coordinated control of dual roles at the cell surface and in the nucleus of these receptors could be instrumental for tumorigenesis. We exemplified here that nuclear TSPAN8 could be detected in multiple human cancers, and the nuclear TSPAN8 is an independent indicator for more aggressive features, advanced stage, and poor clinical outcomes. The transport of TSPAN8 from the cell membrane to the cytoplasm has been linked to endocytic internalization and endosomal sorting. However, the specific cellular context and machinery regulating the nuclear translocation of TSPAN8 from the cytoplasm remain unclear. In our attempts to determine the mechanism underlying TSPAN8 nuclear localization, we found that EGF treatment induced phosphorylation of TSPAN8 at S129 by AKT. Together with our previous study,27 our current work support a model in which EGF–EGFR signaling-activated AKT phosphorylates TSPAN8 to promote its interaction with 14-3-3θ and importin ß1, transporting TSPAN8 into the nucleus where it interacts with STAT3 to regulate target gene transcription (Fig. 9). Consistent with this model, previous studies have shown that nuclear pore complexes (NPCs) and importin ß are involved in the EGFR, ErbB-2 and FGF receptor-1 (FGFR-1) trafficking from the cell surface to the nucleus.54,55,56,57,58 In this model, phosphorylation of TSPAN8 at S129 plays a crucial role in its nuclear transport. In this regard, the TSPAN8S129A mutation that blocks the phosphorylation of S129 not only abrogated the nuclear translocation of TSPAN8, but also decreased proliferation, colony formation and migration ability of cancer cells to a certain extent. In contrast, TSPAN8S129D mutation that mimics constitutive phosphorylation of S129 strongly promoted the nuclear translocation of TSPAN8, as well as the aggressive phenotypes of cancer cells. More importantly, the levels of pTSPAN8S129 were significantly increased in a variety of human malignant tumors including BrCa, and such elevated phosphorylation was positively associated with both nuclear levels of TSPAN8 and aggressive features including mesenchymal phenotype, advanced tumor size, TNBC subtype and distant metastasis. In particular, the levels of pTSPAN8S129 were reversely correlated with overall survival of cancer patients. The IHC scores of TSPAN8 expression, pTSPAN8S129 or nuclear TSPAN8 expression were independent predictors of dismal patient survival time, after adjusting for the relevant clinical covariate in a Cox multivariate model.
A schematic model showing that AKT phosphorylates TSPAN8 at Ser129 upon EGF treatment, resulting in the nuclear translocation of TSPAN8 with the help of 14-3-3θ and importin β1. In nucleus, phosphorylated TSPAN8 interacts with STAT3 to form a complex on chromatin to co-regulate target gene expression and promote tumorigenesis (left part). To target the nuclear TSPAN8-mdiated tumor development, a humanized monoclonal antibody hT8Ab4 targeting the large extracellular loop of TSPAN8 (TSPAN8-LEL) was developed, which can block TSPAN8 cytosol-to-nucleus translocation and thus suppress downstream STAT3-dependent transcriptional activity, displaying an antitumor effect (right part).
To investigate the function of TSPAN8 in the nucleus and the underlying mechanisms, we focused on STAT3, a transcription factor that interacts with phosphorylatedTSPAN8S129. Combined analysis of RNA-Seq and CHIP-Seq revealed that TSPAN8 with S129 phosphorylation enters the nucleus and associates with STAT3 genome-wide to regulate the transcription of downstream target genes including BCL2, MYC, MMP9 etc., which are associated with stemness,59 EMT,60,61 PI3K/AKT,62 TGF-ß63 related pathways. In addition, we further determined the clinical relevance of the TSPAN8–STAT3 interaction, and observed positive correlations among the levels of pTSPAN8S129, nuclear TSPAN8 and nuclear STAT3 in BrCa specimens. It is worth noting that high levels of both pTSPAN8S129 and STAT3 showed better correlation with poor prognosis of breast cancer patients than did pTSPAN8S129 or STAT3 alone.
The EGF–EGFR pathway is widely perceived as one of the most dysregulated signaling pathways in human cancers. Despite tremendous efforts have been made in the development of clinical drug targeting EGFR over the past several decades, the current targeted therapy designed to circumvent EGFR has yielded only modest effects on most cancer types,64,65,66,67 with the exception of those carrying EGFR-activating mutations.68,69,70,71 Even in those cases, most patients eventually developed intrinsic or acquired resistance to EGFR targeted therapy. These frustrating outcomes are, in part, due to the high complicacy and the interactive nature of the EGF–EGFR signaling network. Recently, we and others showed that TSPAN8 play important roles in tumor progression in a variety of human tumors, suggesting that targeting TSPAN8 might be a potential therapeutic strategy.15 Based on the functional and mechanistic understandings of TSPAN8 reported in our current study, we developed a humanized monoclonal antibody specific to TSPAN8. We demonstrated that this anti-TSPAN8 antibody significantly inhibits tumor growth with a dose that did not elicit any detectable side-effects. In this regard, we could not rule out the possibility that the anti-tumor effects of our therapeutic antibody may be partially mediated via blocking functions of TSPAN8 localized on cell membrane. However, the observations that nuclear TSPAN8/pTSPAN8S129 were positively correlated with more aggressive phenotype and dismal prognosis indicate that blocking TSPAN8 nuclear translocation is at least, an important branch of hT8Ab4 efficacy. Thus, our findings provide the first evidence that nuclear TSAPN8 could be a potential therapeutic target in refractory cancer treatment.
It is generally accepted that transmembrane receptors serve as sensors at the cell surface for the recognition of extracellular signals, initiation of ligand-dependent signaling cascade, and/or the generation of second messengers for intracellular signaling.72 Consequently, a range of downstream cytosolic enzymes such as kinases are frequently found to be activated for the nuclear translocation of a diverse array of substrates for various cellular functions.40,41,73 However, recent evidence indicates that membrane receptors can be detected in the nucleus as a full-length form or a fragment of proteolytic cleavage.46 Most of such receptors identified to date belong to RTKs superfamily, including EGFR,47,74,75 insulin-like growth factor 1 receptor (IGF-1R),48,49 fibroblast growth factor receptor (FGFR),76 and vascular endothelial growth factor receptor (VEGFR)77,78,79 etc. Structurally, these RTKs share common characteristics: they are composed of an extracellular domain, a single hydrophobic α-helix transmembrane domain, and an intracellular domain with PTK activity. However, the mechanism by which these cell-surface RTKs travel from cell surface to nucleus remains unclear. The definitive function of these transmembrane proteins in the nucleus has also been overlooked or overshadowed by their traditional function on the cell surface.
The nuclear translocation of membrane proteins with multiple transmembrane domains has been overlooked in the past decades, and the exact molecular mechanism through which these cell-surface proteins travel from the plasma membrane to the nucleus remains unclear. We unearth an instrumental and previously unknown molecular signaling mode, i.e., the activation of AKT by EGF induces TSAPN8 nuclear transport, which has a crucial role in regulating transcription of a series of downstream genes. Our previous study has suggested that site-specific palmitoylation of TSPAN8 is required for binding with cholesterol and nuclear translocation.27 Here, we did not observe an EGF-induced alteration of TSPAN8 palmitoylation. Whether the palmitoylation and cholesterol binding cooperate with EGF–EGFR signaling during the regulation of TSPAN8 nuclear translocation, and the underlying mechanism of how palmitoylation regulate TSPAN8 membrane extraction, warrant further investigation. In summary, our current study helps to advance our knowledge of the nature and role of TSPAN8 nuclear transport in tumorigenesis, provides novel insights into the molecular mechanisms underlying the nuclear translocation of TSPAN8 as well as the function of nuclear TSPAN8, and presents a new therapeutic strategy for BrCa.
Materials and methods
Antibodies
All antibodies except the therapeutic hT8Ab4 used in this study were purchased from commercial companies. They are anti-Tspan8 antibody (Abcam, ab70007), anti-GAPDH antibody (Proteintech, 60004-1), anti-Histone H3 antibody (CST, 4499), anti-α-Tubulin antibody (Sigma, T6199), anti-β-actin antibody (Sigma, A2228), anti-Flag antibody (Sigma, M3165), anti-HA antibody (Abcam, ab137838), anti-Myc antibody (Santa Cruz, sc-40) anti-AKT antibody (CST, 9272), anti-pSer/Thr antibody (Abcam, ab17464), anti-STAT3 antibody (CST, 9139), anti-14-3-3θ antibody (CST, 9638), and anti-Ki67 (Abcam, ab15580).
Plasmids
Flag-TSPAN8 and its variants were generated as described previously.27 STAT3 and SLMAP were subcloned into a Myc-tagged vector. All lentiviral plasmids were constructed in a modified pLKO.1 vector.
Cell culture
Briefly, MCF-7 cells, MDA-MB-231 cells, SW1990 cells and HEK293FT cells were obtained from cell line resource of National Infrastructure (Beijing, China) and were grown in DMEM (Invitrogen/Thermo Fisher Scientific, MA, USA) medium supplemented with 10% heat-inactivated FBS (Biological Industries) and 1% penicillin/streptomycin (Gibco/Thermo) at 37 °C with 5% CO2 in a humidified incubator (Thermo). Cells were passaged for ≤ 3 months from the frozen early-passage stocks that had been received from the indicated sources. During the study, all cell cultures were periodically tested for mycoplasma using MycoAlert™ Mycoplasma Detection Kits (Lonza, ME, USA).
Indirect immunofluorescent staining
The indirect immunofluorescent staining assay was performed followed a previous protocol.80 Briefly, Cells were plated on coverslips in six-well plates and were treated with or without indicated dose of EGF (Abbkine, Wuhan, China). Coverslips with the cells were washed once with PBS and fixed in 4% formaldehyde in PBS for 15 min. After permeabilization with Triton X-100 (0.25%) in PBS for 15 min, cells were blocked with PBS containing BSA (5%) for 30 min and then incubated with primary antibodies for 1 h. After three separate washes, cells were incubated with a secondary antibody for another hour and then stained with DAPI for 2 min. The coverslips were washed extensively and fixed on slides. Images were captured using a Leica laser scanning confocal microscope (Leica TCS SP2 AOBS).
Immunoprecipitation and immunoblotting
For whole cell extracts, cells were lysed using Western blotting buffer containing 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% Triton X-100, and 1 mM EDTA supplemented with 1× Cocktail (11873580001, Roche) on shaking bed at 4 °C for 20 min. Then, the lysates were obtained by 4 °C centrifugation at 13,000 rpm for 30 min, the supernatants were boiled with 5× SDS loading buffer. Finally, the proteins were separated by SDS-PAGE, transferred to a PVDF membrane (Bio-Rad), and blotted with indicated antibodies. Western blotting images were captured by the Mini Chemiluminescent Imaging and Analysis System (Beijing Sage Creation Science Co., Ltd.).
For immunoprecipitation assay, cells lysates were obtained using immunoprecipitation buffer (IP buffer) containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5% NP-40, and 1 mM EDTA supplemented with Cocktail. After centrifugation, the supernatants were incubated with Flag-beads (GenScript) or indicated antibodies together with protein A/G beads (Santa Cruz Biotechnologies) at 4 °C overnight. The beads were then washed with IP buffer for three time and boiled with 1× SDS loading buffer. The interacted proteins were detected using indicated primary antibodies as stated above.
Real-time PCR and RNA-Seq
Total RNAs were extracted from cells using Total RNA Extraction Reagent (R401-01-AA, Vazyme), then cDNAs were reverse-transcribed using the HiScriptII Q Select RT SuperMix (R223-01, Vazyme) with the manufacturer’s guidelines. Real-time PCR was performed using Real-time PCR Master Mix (YEASEN, Shanghai, China) according to an Applied Biosystems Step Two Real-Time PCR System (Applied Biosystems, USA). The primers used were as follows: TSPAN8 forward: 5′-TGCCTGGAGATAGCCTTTGC-3′; TSPAN8 reverse: 5′-ACCACATAGCCAGAACAAGAAG-3′; GAPDH forward: 5′-GGCATCCTGGGCTACACTGA-3′, GAPDH reverse: 5′-GAGTGGGTGTCGCTGTTGAA-3′.
For samples sent for RNA-Seq, we extracted total RNA using the Total RNA Extraction Reagent (R401-01-AA, Vazyme). RNA quality was further assessed on a 2100 expert Bioanalyzer and then total RNAs were sent for library preparation and sequencing on the Illumina Hisseq2000 platform by BGI genomics (Wuhan, China).
Proximity labeling assays
For Mem-TurboID assay, MDA-MB-231 cells were co-transfected with constructs encoding membrane-localized TurboID (TurboID-Myparm) and Flag-TSPAN8 for 36 h. Cells were first treated with biotin for 1 h to induce local protein biotinylation followed by treatment with EGF as indicated. After that, Cells were processed for fractionation assay to collect the cell membrane and nuclear fractions for subsequent streptavidin immunoprecipitation. At last, the presence of biotinylated TSPAN8 in each compartment was examined by western blotting.
For TSPAN8 BioID assay, MDA-MB-231 cells were transfected with TSPAN8-BirA* construct for 48 h. Cells were incubated with medium supplemented with 200 μM Biotin for 12 h (B4639, Sigma) and treated with or without EGF. After that, cells were lysed in NETN buffer at 4 °C for 20 min. After centrifugation, the supernatants were subjected to immunoprecipitation using streptavidin beads (SA021005, Smart-life sciences) for 2 h at 4 °C. Samples were washed with NETN for three times before processed for MS analysis (Majorbio, Shanghai).
Flag pull-down assay
Briefly, proteins including Flag-TSPAN8, Myc-SLMAP, and Myc-STAT3 were purified using the eukaryote cell free system TNT T7 Quick coupled transcription/translation (PR-L1170, Promega). After that, equal amount of indicated proteins together with Flag beads were co-incubated with rotation at 4 °C for 4 h. Samples were washed with IP buffer for three times before addition of 1× SDS sampling buffer. The input and immunoprecipitation samples were finally analyzed via western blotting.
Cell subcellular fractionation assay
Briefly, different cellular compartments of MDA-MB-231 or MCF-7 cells treated as indicated were separated using the commercial Subcellular Protein Fractionation Kit (78840, Thermo Scientific™). After that, the isolated parts were processed for western blotting analysis or Co-IP assay as stated.
ChIP-Seq
ChIP-Seq was performed based on a previous protocol with minor modifications.81,82 Briefly, cells were first cross-linked with freshly prepared formaldehyde (final concentration 1.42%) for 15 min, followed by addition of glycine (125 mM) for 5 min at room temperature. After two-round wash with cold PBS, cells were scraped and collected by centrifugation. Pelleted cells were resuspended in 400 μL ChIP digestion buffer (20 mM Tris-HCl, pH 7.5; 15 mM NaCl; 60 mM KCl; 1 mM CaCl2 and 1× Cocktail inhibitors). To shear chromatin, cells were digested with a proper amount of Micrococcal Nuclease (MNase, M0247S, NEB) at 37 °C for 20 min to ensure that the majority of chromatin was mono- and di-nucleosomes. The reaction was stopped with 2× stop buffer (100 mM Tris-HCl, pH 8.1; 20 mM EDTA; 200 mM NaCl; 2% Triton X-100; 0.2% Na-deoxycholate and protease inhibitors). Samples were further sonicated with Bioruptor at high power for 15 cycles (30-s on/off). After clarification by centrifugation, soluble chromatin was immunoprecipitated with ChIP-grade antibodies against TSPAN8 and STAT3 at 4 °C overnight together with prewashed protein A/G beads.
After an extensive wash with indicated buffers, samples were eluted and reverse cross-linked in elution buffer (10 mM Tris-HCl, pH 8.0; 10 mM EDTA; 150 mM NaCl; 5mM DTT and 1% SDS) at 65 °C overnight. After sequential digestion with DNase and Proteinase K, DNA was purified using the PCR purification kit (B518141, Sangon Biotech). DNAs obtained from three immunoprecipitations after ChIP were pooled to generate libraries with the Ovation Ultra-Low Library Prep kit (NuGEN) according to the manufacturer’s instructions. Sequencing was performed on an Illumina HiSeq 2500 platform (Shbio, Shanghai).
ChIP-qPCR
The cells were fixed and collected following the same procedure as ChIP-Seq. After this, pelleted cells were resuspended in 400 μL ChIP lysis buffer (50 mM Hepes/KOH, pH 7.5; 140 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% Na-deoxycholate and protease inhibitors) and subjected to sonication with Bioruptor to shear the chromatin (high power, 30-s on/off, 20 cycles). After sonication, samples were further diluted twice with lysis buffer and centrifuged to clear the supernatant. An aliquot of 80 μL supernatant (1/10) was directly processed for total DNA extraction as whole cell input. The remaining supernatants were transferred to new Eppendorf tubes and were incubated with either IgG or antibodies described above at 4 °C overnight. Samples were added with prewashed protein A/G beads (L2118; Santa Cruz) for another 3 h. After that, samples were washed five times with indicated buffers and were mixed with 100 μL of 10% chelex (1421253; Bio-Rad). The samples were boiled for 10 min and centrifuged at 4 °C for 1 min. Supernatants were transferred to new tubes. After that, another 120 μL of MilliQ water was added to pellet of each beads. Samples were vortexed for 10s, and were centrifuged again to spin down the beads. The supernatants were combined together as templates for follow-up qPCR analysis. The primers used are shown as follows: BCL2 gene promoter: forward: 5′-TATGTGAGAGAAGTTGGCTTG-3′, reverse: 5′-AGATGGCTTTTGCTATGTTG-3′; MYC gene promoter: forward: 5′-ACTCATTCGTTCGTCCTTC-3′; 5′- CCTCGCTCCACACAATAC-3′; MMP9 gene promoter: forward: 5′-GGAGATTTGGCTGCATGG-3′, reverse: 5′-GCAGGATATGGGGGAAAATAAT-3′.
Xenograft tumor model
Healthy BALB/c nude mice (4-week-old, male) from SLAC Laboratory Animal (Shanghai, China) were maintained in specific pathogen-free conditions in available watered and ventilated cages on a 12-h light/dark cycle in accordance with guidelines of the Institutional Animal Care and Use Committee of the Center for Excellence in Molecular Cell Science (CEMCS), CAS. The approval ID for the use of animals was SIBCB-NAF-14-004-S329-023 issued by the Animal Core Facility of CEMCS.
First, for xenograft tumor formation, around 1 × 106 MCF-7 or MDA-MB-231 cells were injected subcutaneously into BALB/c nude mice. Once tumors were detected, pairs of mice were randomized and intratumorally injected with lentivirus particles with different TSPAN8 constructs (WT or mutants) (MOI = 30) for three successive days. Mice were sacrificed 3 weeks later before being processed for subsequent IHC, RNA scope and tumor volume measurement. Second, for evaluation of therapeutic activity of hT8Ab4 antibody, mice were first injected subcutaneously with MDA-MB-231 cells for 10 days to incubate tumors, then intratumorally injected with therapeutic hT8Ab4 antibody for three successive days. Tumors were monitored at day 0, day 3, day 7, day 14 and day 21 after antibody injection. At last, for evaluation of drug toxicity of hT8Ab4 antibody, BALB/c nude mice were intraperitoneally injected with 50mg/kg or 100mg/kg hT8Ab4 antibody for three successive days, and body weights were measured at day 1, day 5, day 10, day 15, day 20, and day 25.
Human BrCa tissue microarray
All samples used were derived from 95 BrCa patients approved by the ethical committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China, permit number 2019SQ220-1). Detailed clinical information was collected from all patients. The median follow-up period was 73 months (range, 34~104 months). Tissue microarray sections were prepared by Outdo Biotech (Shanghai, China).
Tumor microarray sections were incubated with anti-TSPAN8 antibody (1:50 dilution), anti-phospho-TSPAN8 (S129) antibody (1:25 dilution) or anti-Stat3 antibody (1:100 dilution). The IOD of the immunostaining was also measured by using Image-Pro Plus Version 6.2 software (Media Cybernetics Inc, Bethesda, MD), and the cutoff value for the definition of subgroups was the median IOD. The IOD > median was considered as high expression group, and IOD ≤ median was considered as low expression group.
Isolation and preparation of PDOs
The PDOs were isolated and prepared following our protocol described previously.45 Briefly, the fresh tumor tissue derived from TBNC patients were dismembered into large pieces in a centrifuge tube. The tissues were digested in T25 flasks using SFM (Invitrogen,12634034) medium supplemented with collagenase IV (1:1000) for 45 min in a 37 °C incubator with radiation at 95–100 rpm. After vigorous shaking, the upper layer of white flocculent mucus (mostly collagen) was aspirated and the residual tissues were filtered. The filtered samples were centrifugated at 800 rpm for 10 min to remove the supernatants. The precipitates which contain the cancerous tissue were washed sequentially with 15 mL PBS and 1 mL DMEM/F12 (Invitrogen,12634034) medium, respectively. Then, the mixture of around 40 μL of matrix gel (Corning,356231) and 10 μL of medium was dropped into the six-well plate (50 μL per well). After that, around 600 μL organoid culture medium was injected along the wall for further PDO incubation. The culture medium was composed by 2 mM GlutaMAX™, 10 mM HEPS, 100 U/mL Penicillin-Streptomycin, 1.25 mM N-acetylcysteine, 5 ng/mL EGF, 0.5 nmol/L A83-01, 5 µM Y-27632, 500 nM SB202190, B-27(1:50), Recombinant Human Noggin Protein (1:50) and R-spondin-1 (1:20).
For PDO passage and transmission, the organoids were transferred into a 50 mL centrifuge tube and centrifuged at 900 rpm for 5 min to remove the supernatants and matrix gel. The PDOs were first digested with Triple enzyme (Invitrogen,12605036) for 5 min at 37 °C into individual cells, after that, serum-containing medium was added in a ratio of 1:1 with triple enzyme to terminate the digestion before subjecting to centrifuge at 400 rpm for 5 min. The cells were resuspended with mixture of matrix gel and medium (4:1) and subdivided into 24-well plate for further passage or experimental testing.
Statistical analysis
Statistical analysis was performed using the SAS statistical software package (9.1.3). Quantification data are presented as means ± SD from three independent experiments unless otherwise stated. Continuous data were compared using unpaired Student’s t-test or paired t-test (comparison between two variables) or one-way ANOVA analysis with Bonferroni post hoc test (comparison of multiple variables). Survival curves were calculated according to the Kaplan–Meier method. P < 0.05 were considered statistically significant.
References
Zeng, A. et al. Prospectively isolated tetraspanin(+) neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173, 1593–1608.e20 (2018).
Zimmerman, B. et al. Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167, 1041–1051.e11 (2016).
Yang, Y. et al. Open conformation of tetraspanins shapes interaction partner networks on cell membranes. EMBO J. 39, e105246 (2020).
Huang, Y. et al. Migrasome formation is mediated by assembly of micron-scale tetraspanin macrodomains. Nat. Cell Biol. 21, 991–1002 (2019).
Yanez-Mo, M., Barreiro, O., Gordon-Alonso, M., Sala-Valdes, M. & Sanchez-Madrid, F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 (2009).
Liu, Z. et al. Tetraspanins TSP-12 and TSP-14 function redundantly to regulate the trafficking of the type II BMP receptor in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 117, 2968–2977 (2020).
Nankivell, P. et al. Tetraspanins CD9 and CD151, epidermal growth factor receptor and cyclooxygenase-2 expression predict malignant progression in oral epithelial dysplasia. Br. J. Cancer 109, 2864–2874 (2013).
Liu, Z. et al. Promotion of bone morphogenetic protein signaling by tetraspanins and glycosphingolipids. PLoS Genet. 11, e1005221 (2015).
Levy, S. & Shoham, T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5, 136–148 (2005).
Hemler, M. E. Targeting of tetraspanin proteins-potential benefits and strategies. Nat. Rev. Drug Discov. 7, 747–758 (2008).
Hemler, M. E. Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 14, 49–60 (2014).
Wu, D. et al. Pairing of integrins with ECM proteins determines migrasome formation. Cell Res. 27, 1397–1400 (2017).
Zhang, S., Kodys, K., Babcock, G. J. & Szabo, G. CD81/CD9 tetraspanins aid plasmacytoid dendritic cells in recognition of hepatitis C virus-infected cells and induction of interferon-alpha. Hepatology 58, 940–949 (2013).
Tomlinson, M. G. Eye-opening potential for tetraspanin Tspan12 as a therapeutic target for diseases of the retinal vasculature. Circulation 136, 196–199 (2017).
Zoller, M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9, 40–55 (2009).
McAllister, S. S. & Weinberg, R. A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 16, 717–727 (2014).
Greco, C. et al. E-cadherin/p120-catenin and tetraspanin Co-029 cooperate for cell motility control in human colon carcinoma. Cancer Res. 70, 7674–7683 (2010).
Berthier-Vergnes, O. et al. Gene expression profiles of human melanoma cells with different invasive potential reveal TSPAN8 as a novel mediator of invasion. Br. J. Cancer 104, 155–165 (2011).
Zhang, H. S., Liu, H. Y., Zhou, Z., Sun, H. L. & Liu, M. Y. TSPAN8 promotes colorectal cancer cell growth and migration in LSD1-dependent manner. Life Sci. 241, 117114 (2020).
Cheng, I. et al. Type 2 diabetes risk variants and colorectal cancer risk: the Multiethnic Cohort and PAGE studies. Gut 60, 1703–1711 (2011).
Schafer, D. et al. Identification of CD318, TSPAN8 and CD66c as target candidates for CAR T cell based immunotherapy of pancreatic adenocarcinoma. Nat. Commun. 12, 1453 (2021).
Liu, Y. et al. Extracellular vesicle tetraspanin-8 level predicts distant metastasis in non-small cell lung cancer after concurrent chemoradiation. Sci. Adv. 6, eaaz6162 (2020).
Zhu, R. et al. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat. Commun. 10, 2863 (2019).
Fu, N. Y. et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 19, 164–176 (2017).
Wang, H., Rana, S., Giese, N., Buchler, M. W. & Zoller, M. Tspan8, CD44v6 and alpha6beta4 are biomarkers of migrating pancreatic cancer-initiating cells. Int. J. Cancer 133, 416–426 (2013).
Li, J. et al. SOX9 is a critical regulator of TSPAN8-mediated metastasis in pancreatic cancer. Oncogene 40, 4884–4893 (2021).
Huang, Y. et al. Nuclear translocation of the 4-pass transmembrane protein Tspan8. Cell Res. 31, 1218–1221 (2021).
Schmidt, F. et al. Tetraspanin 8 is an interactor of the metalloprotease meprin beta within tetraspanin-enriched microdomains. Biol. Chem. 397, 857–869 (2016).
Yang, Y. C. et al. Circulating proteoglycan endocan mediates EGFR-driven progression of non-small cell lung cancer. Cancer Res. 80, 3292–3304 (2020).
Pan, M. et al. EpCAM ectodomain EpEX is a ligand of EGFR that counteracts EGF-mediated epithelial-mesenchymal transition through modulation of phospho-ERK1/2 in head and neck cancers. PLoS Biol. 16, e2006624 (2018).
Claperon, A. et al. Hepatic myofibroblasts promote the progression of human cholangiocarcinoma through activation of epidermal growth factor receptor. Hepatology 58, 2001–2011 (2013).
Ponsioen, B. et al. Quantifying single-cell ERK dynamics in colorectal cancer organoids reveals EGFR as an amplifier of oncogenic MAPK pathway signalling. Nat. Cell Biol. 23, 377–390 (2021).
Gao, M. et al. EGFR activates a TAZ-driven oncogenic program in glioblastoma. Cancer Res. 81, 3580–3592 (2021).
Luo, H. et al. ALCAM-EGFR interaction regulates myelomagenesis. Blood Adv. 5, 5269–5282 (2021).
Sigismund, S. et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA 102, 2760–2765 (2005).
Akdag, M. et al. Proximal biotinylation-based combinatory approach for isolating integral plasma membrane proteins. J. Proteome Res. 19, 3583–3592 (2020).
Wei, W. et al. Cell type-selective secretome profiling in vivo. Nat. Chem. Biol. 17, 326–334 (2021).
Bogdan, S. & Klambt, C. Epidermal growth factor receptor signaling. Curr. Biol. 11, R292–R295 (2001).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell. Biol. 196, 801–810 (2012).
Pekarsky, Y. et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc. Natl. Acad. Sci. USA 97, 3028–3033 (2000).
Manning, B. D. & Cantley, L. C. AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 (2007).
Rouillard, A. D. et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 (2016).
Iwamaru, A. et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 26, 2435–2444 (2007).
Ren, X. et al. Identification of Niclosamide as a New Small-Molecule Inhibitor of the STAT3 Signaling Pathway. ACS Med. Chem. Lett. 1, 454–459 (2010).
Zhou, T. H. et al. Establishment and optimization of co-culture technology for breast cancer organoids. J. Shanghai Jiao Tong Univ. (Med. Sci.) 41, 1017–1024 (2021).
Sardi, S. P., Murtie, J., Koirala, S., Patten, B. A. & Corfas, G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 127, 185–197 (2006).
Wang, S. C. & Hung, M. C. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 15, 6484–6489 (2009).
Aleksic, T. et al. Nuclear IGF1R interacts with regulatory regions of chromatin to promote RNA polymerase II recruitment and gene expression associated with advanced tumor stage. Cancer Res. 78, 3497–3509 (2018).
Asmane, I. et al. Insulin-like growth factor type 1 receptor (IGF-1R) exclusive nuclear staining: a predictive biomarker for IGF-1R monoclonal antibody (Ab) therapy in sarcomas. Eur. J. Cancer 48, 3027–3035 (2012).
Liccardi, G., Hartley, J. A. & Hochhauser, D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 71, 1103–1114 (2011).
Lo, H. W. et al. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 65, 338–348 (2005).
Lo, H. W. & Hung, M. C. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br. J. Cancer 94, 184–188 (2006).
Psyrri, A. et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin. Cancer Res. 11, 5856–5862 (2005).
Reilly, J. F. & Maher, P. A. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J. Cell Biol. 152, 1307–1312 (2001).
Giri, D. K. et al. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol. Cell Biol. 25, 11005–11018 (2005).
Wang, Y. N. et al. The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. Chem. 285, 38720–38729 (2010).
Stachowiak, M. K., Maher, P. A. & Stachowiak, E. K. Integrative nuclear signaling in cell development-a role for FGF receptor-1. DNA Cell Biol. 26, 811–826 (2007).
Liao, H. J. & Carpenter, G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol. Biol. Cell 18, 1064–1072 (2007).
He, W. et al. IL22RA1/STAT3 signaling promotes stemness and tumorigenicity in pancreatic cancer. Cancer Res. 78, 3293–3305 (2018).
Rokavec, M. et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 124, 1853–1867 (2014).
Lin, W. H. et al. STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT-MET switch and cancer metastasis. Oncogene 40, 791–805 (2021).
Niit, M. et al. Cell-cell and cell-matrix adhesion in survival and metastasis: Stat3 versus Akt. Biomol. Concepts. 6, 383–399 (2015).
Dees, C. et al. TGF-beta-induced epigenetic deregulation of SOCS3 facilitates STAT3 signaling to promote fibrosis. J. Clin. Invest. 130, 2347–2363 (2020).
Green, M. D. et al. Gefitinib treatment in hormone-resistant and hormone receptor-negative advanced breast cancer. Ann. Oncol. 20, 1813–1817 (2009).
Dickler, M. N. et al. A phase II trial of erlotinib in combination with bevacizumab in patients with metastatic breast cancer. Clin. Cancer Res. 14, 7878–7883 (2008).
Thomas, M. B. et al. Phase II trial of the combination of bevacizumab and erlotinib in patients who have advanced hepatocellular carcinoma. J. Clin. Oncol. 27, 843–850 (2009).
von Minckwitz, G. et al. A multicentre phase II study on gefitinib in taxane- and anthracycline-pretreated metastatic breast cancer. Breast Cancer Res. Treat. 89, 165–172 (2005).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
van den Bent, M. J. et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J. Clin. Oncol. 27, 1268–1274 (2009).
Reardon, D. A. et al. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J. Neurooncol. 96, 219–230 (2010).
Brand, T. M., Iida, M. & Wheeler, D. L. Molecular mechanisms of resistance to the EGFR monoclonal antibody cetuximab. Cancer Biol. Ther. 11, 777–792 (2011).
Case, L. B., Ditlev, J. A. & Rosen, M. K. Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 48, 465–494 (2019).
Arkhipov, A. et al. Architecture and membrane interactions of the EGF receptor. Cell 152, 557–569 (2013).
Chen, D. J. & Nirodi, C. S. The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin Cancer Res. 13, 6555–6560 (2007).
Chen, H., Chen, X. & Zheng, Y. The nuclear lamina regulates germline stem cell niche organization via modulation of EGFR signaling. Cell Stem Cell 13, 73–86 (2013).
Bryant, D. M. & Stow, J. L. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic 6, 947–954 (2005).
Wells, A. & Marti, U. Signalling shortcuts: cell-surface receptors in the nucleus? Nat. Rev. Mol. Cell Biol. 3, 697–702 (2002).
Massie, C. & Mills, I. G. The developing role of receptors and adaptors. Nat. Rev. Cancer 6, 403–409 (2006).
Wang, S. C. et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8, 1359–1368 (2006).
An, L. et al. Dual-utility NLS drives RNF169-dependent DNA damage responses. Proc. Natl. Acad. Sci. USA 114, E2872–E2881 (2017).
Wal, M. & Pugh, B. F. Genome-wide mapping of nucleosome positions in yeast using high-resolution MNase ChIP-Seq. Methods Enzymol. 513, 233–250 (2012).
Meng, Y. et al. A TNFR2-hnRNPK Axis Promotes Primary Liver Cancer Development via Activation of YAP Signaling in Hepatic Progenitor Cells. Cancer Res. 81, 3036–3050 (2021).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82073269, 81772802, M-0349, 32070710, 81902806 and 82002820), Shanghai Science and Technology Innovation Action Plan (20XD1402800), Clinical Research Plan of SHDC (SHDC2020CR2065B), Clinical Research Innovation Plan of Shanghai General Hospital (CTCCR-2016B05), the Shanghai Pujiang Program (19PJ1408300), the Open Research Fund of State Key Laboratory of Genetic Engineering, Fudan University (SKLGE-2103), Natural Science Foundation of Jiangsu Province (BK20181186) and Shanghai Science and Technology Commission (20JC1410100).
Author information
Authors and Affiliations
Contributions
H.W., X.L. and L.A. designed research. X.L., L.A., G.F. and L.Z performed most cellular and in vitro experiments with input from W.H., J.Li., and Y.C.; J.Liu., W.G., J.X. G.F., S.D, T.Z. and H.Y carried out the PDO assay. Y.H. and L.Y. helped imaging analysis and quantification. L.A. and S.J. analyzed the IHC data. All authors discussed and commented on the manuscript. H.W., L.A. and G.F. wrote and revised the paper. H.W. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Lu, X., An, L., Fan, G. et al. EGFR signaling promotes nuclear translocation of plasma membrane protein TSPAN8 to enhance tumor progression via STAT3-mediated transcription. Cell Res 32, 359–374 (2022). https://doi.org/10.1038/s41422-022-00628-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-022-00628-8
This article is cited by
-
IRG1/Itaconate inhibits proliferation and promotes apoptosis of CD69+CD103+CD8+ tissue-resident memory T cells in autoimmune hepatitis by regulating the JAK3/STAT3/P53 signalling pathway
Apoptosis (2024)
-
The versatile roles of testrapanins in cancer from intracellular signaling to cell–cell communication: cell membrane proteins without ligands
Cell & Bioscience (2023)
-
Tetraspanins predict the prognosis and characterize the tumor immune microenvironment of glioblastoma
Scientific Reports (2023)
-
TSPAN8 regulates EGFR/AKT pathway to enhance metastasis in gastric cancer
Molecular Biology Reports (2023)
-
Nuclear tetraspanin 8 promotes breast cancer progression
Cell Research (2022)