Abstract
Severe COVID-19 disease caused by SARS-CoV-2 is frequently accompanied by dysfunction of the lungs and extrapulmonary organs. However, the organotropism of SARS-CoV-2 and the port of virus entry for systemic dissemination remain largely unknown. We profiled 26 COVID-19 autopsy cases from four cohorts in Wuhan, China, and determined the systemic distribution of SARS-CoV-2. SARS-CoV-2 was detected in the lungs and multiple extrapulmonary organs of critically ill COVID-19 patients up to 67 days after symptom onset. Based on organotropism and pathological features of the patients, COVID-19 was divided into viral intrapulmonary and systemic subtypes. In patients with systemic viral distribution, SARS-CoV-2 was detected in monocytes, macrophages, and vascular endothelia at blood–air barrier, blood–testis barrier, and filtration barrier. Critically ill patients with long disease duration showed decreased pulmonary cell proliferation, reduced viral RNA, and marked fibrosis in the lungs. Permanent SARS-CoV-2 presence and tissue injuries in the lungs and extrapulmonary organs suggest direct viral invasion as a mechanism of pathogenicity in critically ill patients. SARS-CoV-2 may hijack monocytes, macrophages, and vascular endothelia at physiological barriers as the ports of entry for systemic dissemination. Our study thus delineates systemic pathological features of SARS-CoV-2 infection, which sheds light on the development of novel COVID-19 treatment.
Similar content being viewed by others
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel beta-coronavirus, has given rise to a global pandemic. In comparison with the other two coronavirus strains responsible for Middle Eastern respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS), SARS-CoV-2 is associated with lower mortality but with high transmission efficiency.1 A large body of clinical data from tertiary referral centers showed that although most patients with COVID-19 show no symptoms or mild symptoms, the elders with underlying diseases are prone to developing severe or critical disease condition with high mortality.2,3,4 In addition to respiratory manifestations, extra-pulmonary involvements are increasingly recognized, including dysfunctions of the immune system, cardiovascular system, gastrointestinal system, and neurological system.3,4 However, regarding the pathogenicity of COVID-19, it remains unclear whether the systemic pathological changes are driven by local viral attack, the maladaptive immune responses, or the consequences of therapeutics.
Autopsies of COVID-19 deceased patients and experimental animal studies have provided insights into the fundamental pathological changes in the lungs and multiple extrapulmonary organs.5,6,7,8,9,10,11,12 We and others have found that the lungs are the most affected organ by SARS-CoV-2, showing diffuse alveolar damage, exudation, interstitial fibrosis, extensive infiltration of immune cells, including dysfunctional alveolar macrophages, and abundant inflammatory factors.7,13,14 Extrapulmonary organs exhibited different extent of tissue injuries and inflammatory responses. In particular, the lymphatic organs such as the spleen and lymph nodes contained reduced lymphocytes but increased macrophages.7 These findings provide the pathological basis for severe hypoxia (acute respiratory distress syndrome (ARDS)) and immune malfunction in critically ill patients with COVID-19. However, information about the tissue tropism of SARS-CoV-2 and the port of virus entry responsible for systemic dissemination is sparse. A recent study provides evidence for SARS-CoV-2 entry into the nervous system by crossing the neural-mucosal interface in olfactory mucosa.15 However, it remains unknown whether SARS-CoV-2 penetrates through physical barriers into target organs.
It is believed that definitive detection of SARS-CoV-2 spike or nucleocapsid protein by immunohistochemical (IHC) staining should confirm in situ viral presence in the lungs and extrapulmonary organs, thereby establishing direct evidence of viral infection of target organs.16,17,18,19 In this study, we profiled SARS-CoV-2 organotropism using 26 autopsy cases from four cohorts in Wuhan, China, and evaluated virus-associated organ injuries. We classified COVID-19 into two pathological subtypes as intrapulmonary infection and systemic infection respectively. Our study delineates SARS-CoV-2 organotropism and pathogenicity, which provides a better understanding of the mechanisms of viral infection and novel therapeutic approaches to minimizing the systemic dissemination of the virus.
Results
Characteristics of the cohort
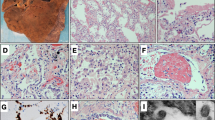
All 26 autopsy cases used in this study were deceased patients (median 67.5-year-old) and met the diagnostic criteria for critically ill status of COVID-19. The presence of SARS-CoV-2 in all cases was confirmed by PCR tests, IHC staining of viral spike or nucleoprotein, or electron microscopy. The basic information, disease duration, clinical manifestations, and treatment regimens for 26 cadaver donors were summarized in Tables 1, 2; Supplementary information, Fig. S1. Twenty cases (76.9%) were diagnosed with underlying diseases, including chronic pulmonary diseases (23.1%), chronic cardiovascular diseases (38.5%), and hypertension (34.6%). The median duration from COVID-19 symptom onset to death was 38.5 days. The major death causes for our COVID-19 autopsy cases included severe pulmonary injuries (COVID-19-related respiratory failure with/without pulmonary fungal infection), pulmonary thromboembolism, dissecting aneurysm rupture, and cardiovascular disorders (Fig. 1a and Table 1).
a Major death causes for the 26 autopsy COVID-19 cases. The major death causes were severe pulmonary injuries (n = 23), including COVID-19-related respiratory failure without (n = 21) or with (n = 2) pulmonary fungal infection. The major death causes for other three cases were pulmonary thromboembolism, dissecting aneurysm rupture, and cardiovascular disorders, respectively. b Schematic model for SARS-CoV-2 organ tropism. LNs, Lymph nodes. c Heatmap showing SARS-CoV-2 distribution groups and viral RNA (Log2) in postmortem organs in 26 autopsy cases with COVID-19. LU, left upper; LL, left lower; RU, right upper; RM; right middle; RL, right lower. d Percentage of COVID-19 autopsy cases in three groups of SARS-CoV-2 distribution. e The correlation between SARS-CoV-2 viral RNA in the lungs and the number of SARS-CoV-2-positive organs. f Comparison of viral infection rate between SARS-CoV-2 based on the current autopsy study and SARS-CoV in the literature in postmortem organs from patients with COVID-19 and SARS.
Overview of SARS-CoV-2 virus organotropism
We and others have identified the pathological changes of multiple organs through systemic autopsy examination of COVID-19 patients,5,6,7,8,9,10 although information about the duration of SARS-CoV-2 in target organs and its correlation with disease progression is sparse. Through systemic autopsy examination, we found that SARS-CoV-2 RNA, spike protein or virion-like particles existed in the lungs and multiple extrapulmonary organs in critically ill patients as long as 15–67 days after symptom onset. The SARS-CoV-2 viral RNA distributed in postmortem organs including those in the respiratory, digestive, genitourinary, cardiovascular, immune systems, endo/exocrine glands, and skin (Fig. 1b, c). The viral distribution patterns were categorized into three groups: systemic (12/26, 46.2%), intrapulmonary (12/26, 46.2%), and others (2/26, 7.7%) (Fig. 1b–d). Although COVID-19-related respiratory failure was present in both systemic and intrapulmonary distribution groups, cases in the systemic distribution group also exhibited increased probabilities of extrapulmonary organ failures (especially the failure of kidney, heart, and liver). Two cases (Cases 25 and 26) categorized into other distribution group did not show SARS-CoV-2 RNA in the lungs at the time of death, but they manifested positive SARS-CoV-2 RNA signal in the digestive tract or skin. Intriguingly, these two patients did not show typical COVID-19 pneumonia but died of heart failure caused by dilated cardiomyopathy and infective endocarditis (Case 25) or respiratory failure caused by severe Aspergillus infection (Case 26). Notably, the number of SARS-CoV-2-positive organs was correlated with viral RNA in the lungs (Fig. 1e), suggesting that higher pulmonary viral RNA or deficiency in viral clearance may contribute to the broad virus dissemination. To compare virus-tropic organs between COVID-19 and SARS, we reviewed previous literature regarding virus organotropism.20,21,22,23,24,25,26,27,28 A broader spectrum of organ infection was shown by SARS-CoV-2 as compared to SARS-CoV (Fig. 1f). SARS-CoV-2, but not SARS-CoV, was detected in the aorta, esophagus, thyroid gland, testes and uterus, showing a diffused map of SARS-CoV-2 organotropism in critically ill patients with COVID-19.
SARS-CoV-2-associated pulmonary pathology
Pulmonary pathologies in this cohort (including 6 reported cases from Wuhan Jinyintan Hospital7) were featured by three phases of diffuse alveolar damage (DAD): exudation, proliferation, and fibrosis (Fig. 2a; Supplementary information, Fig. S2a).29 DAD-exudation phase was presented with pneumocyte injury and exfoliation, serous or fibrinous exudates, and hyaline membrane formation. DAD-proliferation phase was featured by pneumocyte hyperplasia and atypia, as well as macrophage infiltration. DAD-fibrosis phase manifested alveolar fibrosis and/or interstitial fibrosis. SARS-CoV-2-associated pulmonary pathology was quantified in the first 15 consecutive autopsy cases which were tested with SARS-CoV-2 RNA in the lungs. The percentage of DAD-exudation, DAD-proliferation, and DAD-fibrosis areas accounted for 41.5 ± 6.3%, 16.5 ± 6.3%, and 17.0 ± 7.5%, respectively, of total pulmonary tissues in 15 autopsy cases (Fig. 2b–d). Intriguingly, DAD-fibrosis was found as early as 15 days since the symptom onset and increased with disease progression (Fig. 2d), indicating fibrosis as an early and severe COVID-19 complication. Pulmonary areas with higher expression of SARS-CoV-2 spike protein were featured by hyperproliferation of epithelia (Supplementary information, Fig. S2b, c). Further analyses revealed that the proliferative cells containing SARS-CoV-2 were mainly ACE2-expressing and TTF-1-positive alveolar epithelia and bronchiolar basal cells (Supplementary information, Fig. S2d, e). PCR analyses indicated that pulmonary viral RNA maintained a high level from 15–36 days since the symptom onset and showed a gradual decrease with disease duration extension (36–52 days) (Fig. 2e). We also observed extensive pulmonary changes in all postmortem patients with respiratory failure, including alveolar serous and fibrin exudates, hyaline membrane formation, and alveolar/interstitial fibrosis (Fig. 2f; Supplementary information, Fig. S2f), which may form a histopathological basis for impeded air diffusion. Remarkably, in ten COVID-19 patients with medical records of respiratory failure, mechanical ventilation, and arterial oxygen partial pressure (PaO2), mucus plugs were present in the alveoli or bronchioles (Fig. 2g), which were inversely associated with the levels of PaO2 (Fig. 2h), suggesting that mucus production was increased in hypoxemia and may limit the efficacy of mechanical ventilation during COVID-19 treatment.
a H&E and IHC staining showing SARS-CoV-2 spike protein in pulmonary areas manifesting different features (1, exudation; 2, proliferation; 3, fibrosis) of diffuse alveolar damage (DAD). Scale bars, 250 μm. b–e Proportion of DAD-exudation areas (b), DAD-proliferation areas (c), and DAD-fibrosis areas (d), and the average SARS-CoV-2 RNA (e) in postmortem lungs from 15 COVID-19 autopsy cases. f, g H&E staining showing hyaline membrane formation (f) and bronchiolar-alveolar mucus (g). Scale bars, 100 μm. h The correlation between average bronchiolar-alveolar mucus plug number and PaO2 level in patients with respiratory failure.
Identification of SARS-CoV-2 in the endothelia of physiological barriers in the lungs, kidneys and testes
We found that SARS-CoV-2 spike protein was present in CD34+ endothelia at blood–air barrier or pulmonary vessels in serial sections of the COVID-19 lungs (Fig. 3a), raising the possibility that SARS-CoV-2 was able to infiltrate blood–air barrier for intrapulmonary and systemic dissemination. SARS-CoV-2 spike protein was mainly detected in the glomeruli with abundant endothelium-formed filtration barriers and renal proximal convoluted tubular epithelia in the kidneys (Cases 1, 2, 4, and 5) which were positive for SARS-CoV-2 RNA (Fig. 3b), but was not detected in the kidneys (including Case 16) negative for SARS-CoV-2 RNA (Fig. 3b). SARS-CoV-2 spike protein was also detected in endothelia of the blood–testis barrier (Fig. 3c), spermatogenic cells and stromal cells in the seminiferous tubules, and sperms in the epididymis in the COVID-19 testes positive for SARS-CoV-2 RNA (Cases 2, 5 and 11) (Fig. 3c). These results provide evidence of SARS-CoV-2 presence in the endothelia including those in physiological barriers (blood–air, blood–testis, and filtration barriers), implying that these cells are susceptible to SARS-CoV-2 infection followed by systemic dissemination.
a Immunofluorescent staining of SARS-CoV-2 spike protein and CD34 in endothelia of pulmonary vessels using COVID-19 lung tissues (upper panel, Case 1) or control lung tissues from a patient with lung carcinoma (lower panel). Scale bars, 25 μm. b IHC showing that SARS-CoV-2 spike protein was detected in glomeruli with abundant filtrated barriers and convoluted tubular epithelia in the kidneys positive for viral RNA (Case 2). The kidney tissues (Case 16) negative for viral RNA were used as control. Scale bars, 25 μm. c H&E staining and IHC staining showing SARS-CoV-2 spike in endothelia of the blood–testis barrier, seminiferous tubules, and sperms in the epididymis (blue arrows) of the testes from COVID-19 patients (Case 2). Scale bars, 50 μm.
Identification of SARS-CoV-2 in monocytes and macrophages
To investigate the possible routes for viral dissemination, we investigated whether SARS-CoV-2 was contained in monocytes and macrophages, critical immune cell populations.30 We found that the cellular components of alveolar exudate were mainly CD68+ macrophages positive for SARS-CoV-2 spike protein (Fig. 4a). IHC staining using serial sections also identified the presence of SARS-CoV-2 spike protein in monocytes and macrophages in lymph nodes and the spleen (Fig. 4b, c), as well as peripheral blood mononuclear cells in the postmortem lungs, kidneys, lymph nodes, spleen, and intestines (Fig. 4d). Single-cell RNA-sequencing (scRNA-seq) of lung tissues from a COVID-19 autopsy case (Case 17) within 2 h after death revealed the presence of CD14+ monocytes (CD14+ Mono-1, -2), monocyte-derived alveolar macrophages (MoAM-1, -2), and other cell types (Fig. 4e; Supplementary information, Fig. S3). CD14+ monocytes were characterized by VCAN expression and MoAMs were positive for C1QA and C1QC. Importantly, we detected SARS-CoV-2 transcripts of open reading frame 10 (ORF_10) and nucleocapsid in alveolar CD14+ monocyte (Fig. 4f), confirming the presence of SARS-CoV-2 in lung monocytes. To address whether the entry of SARS-CoV-2 into monocytes and macrophages was mediated by receptors other than ACE2, we measured mRNA of previously reported SARS-CoV-2 receptors including BSG (encoding CD147), TFRC (encoding transferrin receptor-1), and NRP1 (encoding neuropilin-1) in CD14+ monocytes and MoAMs in COVID-19 lungs using scRNA-seq.31,32,33,34 We found that CD14+ Mono-1, -2, and MoAM-1, -2 expressed BSG, TFRC, and NRP1, but not ACE2 (Fig. 4g), suggesting that CD147, transferrin receptor-1, or neuropilin-1 might mediate SARS-CoV-2 infection of monocytes and macrophages. These findings suggest that SARS-CoV-2 may hijack monocytes and macrophages for systemic dissemination.
a IHC staining of CD68, CK7, and viral spike in alveoli on serial sections. Macrophages are indicated by blue arrows. Scale bars, 50 μm. b, c IHC staining of monocytes/macrophages marked by CD68 and viral spike protein in lymph nodes (b) and the spleen (c) on serial sections from COVID-19 patients. Scale bars, 50 μm. d IHC staining showing viral spike in peripheral blood mononuclear cells (blue arrows) in vessels of the indicated postmortem organs from COVID-19 patients. Scale bar, 50 μm. e U-MAP showing scRNA-seq of 1437 cells on COVID-19 autopsy lung tissues (Case 17). CD8+ T, CD8+ T cells; CD14+ Mono-1/2, CD14+ monocyte-1/2; MoAM-1/2, monocyte-derived alveolar macrophages-1/2; AT, alveolar epithelial type 1/2 cells; Erythroid-like, erythroid-like and erythroid precursor cells; EC, endothelial cells; Fibro, fibroblast cells; MKI67+, MKI67+cells; Plasma, plasma cells. f Detection of SARS-CoV-2 transcripts. Plot shows SARS-CoV-2 ORF_10 or nucleocapsid (N) genes in CD14+ monocyte-1 from scRNA-seq. g U-MAP showing the expression of BSG (encoding CD147), TFRC (encoding transferrin receptor-1), NRP1 (encoding neuropilin-1), and ACE2 in the scRNA-seq of COVID-19 lung tissues.
Discussion
Investigations of SARS-CoV-2 organotropism, its duration in the target organs and the correlation with disease progression are pivotal for developing effective strategies for the containment of the pandemic. Some studies revealed SARS-CoV-2 distribution in human tissues and viral shedding in body fluids,18,35,36,37,38 but the translational significance remains unclear. Our pathological evidence of SARS-CoV-2 presence in late stage of critically ill patients with COVID-19 suggests that continued usage of antiviral therapy is indispensable throughout the disease course. Pulmonary areas with more SARS-CoV-2 exhibit increased diffused alveolar damage and airway obstruction associated with ventilation dysfunction, supporting the cytotoxic effect of SARS-CoV-2 that causes respiratory failure. We demonstrated that the majority of autopsy cases with systemic virus distribution showed multiple organ failures, supporting the systemic nature of the disease. A minor fraction of patients (5/26) with mild virus-related pulmonary damages died of co-morbidities or secondary infections, suggesting complex causes of death related to COVID-19.
While accumulating data support the presence of SARS-CoV-2 in extrapulmonary organs, how the virus systemically spreads remains an enigma. Our study identified SARS-CoV-2 presence in endothelia located at several physiological barriers (blood–air, filtration, and blood–testis barriers), raising the possibility that the virus might invade these barriers for dissemination. The virus was also found in vascular endothelia of multiple organs, which may cause vasculitis.39 The injured endothelia may initiate vascular dysfunction and subsequently a procoagulant state to induce thrombosis, as well as ischemic or hemorrhagic changes frequently observed in fatal COVID-19 patients.40 Additional cautions should be exercised to treat vulnerable patients with pre-existing diseases associated with high risk of endothelial injury, such as hypertension and diabetes.
Our scRNA-seq of COVID-19 lung tissues, together with previous RNA-seq and tissue staining data, has revealed the presence of SARS-CoV-2 transcripts in CD14+ monocytes and macrophages infiltrating the lungs, the spleen, lymph nodes, and the kidneys.37,41,42 How monocytes and macrophages became infected remains unclear. Recent scRNA-seq analyses of human tissues have revealed that ACE2 and transmembrane protease serine 2 (TMPRSS2) are rarely expressed in immune cells including monocytes and macrophages.37 Our results together with previous studies have revealed that CD147, a recently identified receptor for SARS-CoV-2, was expressed in epithelial cells, lymphocytes, monocytes, and macrophages.31,32 The expression of CD147 correlates with the abundance of viral RNA in alveolar macrophages.37 Since viremia and viral sepsis in COVID-19 patients have been previously described,43,44,45 the presence of SARS-CoV-2 spike or nucleocapsid protein in monocytes and macrophages may be interpreted as: (1) circulating SARS-CoV-2 directly enters peripheral monocytes or tissue-resident macrophages, as for SARS-CoV and MERS-CoV;45,46 (2) circulating monocytes internalize secretory vesicles or cell debris carrying SARS-CoV-2 through endocytosis; (3) tissue-resident macrophages phagocytize virus-infected cells, followed by viral release from lysosome. Also, antibody recognizing the virus may mediate the infection through antibody-dependent enhancement of infection.47 As antibody-based therapeutics including anti-SARS-CoV-2 monoclonal antibody and IL-6 receptor antagonist tocilizumab are under clinical evaluation,48,49 further investigations of the mechanisms underlying SARS-CoV-2 infection in monocytes and macrophages are urgently warranted.
We and previous studies have identified significant infiltrating monocytes and macrophages in COVID-19 lungs, the spleen, lymph nodes, and the kidneys.7,14,50,51 Monocyte and macrophage composition was further characterized by a reduction of tissue-resident alveolar macrophages and an increased abundance of inflammatory monocyte-derived macrophages in critically ill COVID-19 patients.52 Increased population of monocytes and macrophages is associated with type I interferon response dysfunction, which has been implicated in increased severity of SARS and MERS similar to findings in COVID-19.53 The overloaded monocytes and macrophages may directly interact with interferon-γ-secreting T cells, secrete chemoattractants for immune cell recruitment, or produce pro-inflammatory cytokines to induce alveolitis and subsequent pulmonary damages.54,55 Massive pro-inflammatory cytokines released by monocytes and macrophages are associated with respiratory insufficiency and may lead to cytokine storm-associated shock, multiple organ failure, and death in COVID-19 patients.53 Pro-inflammatory macrophages may also phagocytize lymphocytes in the spleen and lymph nodes, thus contributing to lymphopenia in COVID-19 patients.2,30 In addition, a subset of macrophages harboring tissue repair and fibrosis generation signature has been reported in severe COVID-19 patients, extending the potential pathogenicity of infiltrating macrophages to fibrosis.56,57 Therefore, circulating monocytes and tissue infiltrating macrophages are pivotal for systemic and local immune disorders, viral infection, and tissue injuries. Comprehensive evaluations of the signatures of monocytes and macrophages via multimodal single-cell profiling may promote our understanding of the molecular features, activation status, spatial and chronological distribution of monocytes and macrophages in the unique inflammatory milieu of COVID-19, thus benefiting targeted therapeutics.
Methods
Patients, autopsy, and clinical data collection
The autopsy cases were from Huoshenshan Hospital (n = 8), Taikang Tongji Hospital (n = 5), Zhongfaxincheng Hospital (n = 5), and Wuhan Jinyintan Hospital (n = 8), China. We conducted 26 cases of autopsies from cadaver donors who died from Feb 18th to April 4th, 2020, who had been diagnosed with COVID-19, with written consent from patient’s immediate relatives. Lung tissues from a patient who died from sudden heart death or those from a patient with lung carcinima were used as control tissues for comparison with those from the COVID-19 patients. Fresh or formalin-fixed, paraffin-embedded (FFPE) postmortem specimens were used for viral RNA detection, tissue staining, or morphology analyses. SARS-CoV-2 infection of all cadaver donors was confirmed by virologic tests. Retrospective analyses of case history and clinical manifestations were performed through reviewing the electronic medical records, nursing records, laboratory findings and radiological imaging of the cadaver donors. This study was approved by the ethics committee of Huoshenshan Hospital and is in accordance with regulations issued by the National Health Commission of China and the Helsinki Declaration.
Tissue staining and transmission electron microscopy
Hematoxylin and eosin (H&E) staining, IHC, and transmission electron microscopy (TEM) were performed according to the standard procedure as described previously.58,59 IHC was performed by using the strep-tavidin-biotin-peroxidase technique with diaminobenzidine. Immunofluorescent staining of SARS-CoV-2 and CD34 was performed on FFPE lung sections. Heat-induced antigen epitope retrieval in EDTA (pH 9.0) or citrate buffer (pH 6.0) was applied for optimal detection of antigens on FFPE sections. Sections were incubated overnight at 4 °C with primary antibodies as listed in Supplementary information, Table S1. The specificity and reliability of SARS-CoV-2 spike and nucleoprotein antibody have been verified in previously published literatures59,60,61 and were confirmed by immunocytochemistry staining using in vitro cultured Cercopithecus Vero E6 cells (ATCC® CRL-1586™) transfected with SARS-CoV-2 (Supplementary information, Fig. S4). Vero E6 cells without SARS-CoV-2 were used as control. Staining was visualized by Dako REAL™ EnVision™ Detection System (K5007) followed by counterstaining with hematoxylin. The diluent with control IgG antibodies was used as a negative control. Images were captured by using a digital camera (DP73, Olympus) under a light microscope (BX53, Olympus). The pathological lesions and SARS-CoV-2 spike protein were quantified by two pathologists independently. Briefly, three phases of DAD were evaluated according to the histopathological lesion features. The percentage of each phase of DAD was calculated as the ratio of the pulmonary lesion areas showing histopathological changes of each DAD phase versus the total DAD areas in each slide. For each patient, at least 20 randomly selected 100× microscopic fields were calculated. The percentages of SARS-CoV-2-positive cells in type II pneumocytes and bronchiolar epithelia and Ki67-positive pneumocytes and bronchiolar epithelia in the serial section were quantified under 400× magnification of microscopic fields in at least 10 randomly selected areas for each group.
Real-time reverse transcription PCR
The nucleic acids of SARS-CoV-2 were detected by real-time reverse transcription PCR method with a SARS-CoV-2 Nucleic Acid Detection Kit (Sansure Biotech) according to the manufacturer’s instructions. The sequences of primers and probes of SARS-CoV-2 were obtained from National Institute for Viral Disease Control and Prevention (http://nmdc.cn/#/nCoV) which were listed in Supplementary information, Table S2. Relative levels of SARS-CoV-2 RNA in tissues were normalized to Rnase P to adjust for differences in tissue input.
ScRNA-seq and data processing
ScRNA-seq of lung tissues was performed on a COVID-19 autopsy case (Case 17) from whom unfixed lung tissues (right lower lobe presented with focal consolidation) were isolated to single cells within 2 h after tissue collection in a biosafety level-3 (BSL-3) laboratory. Briefly, tissues were digested in GEXSCOPE Tissue Dissociation Solution (Singleron Biotechnologies, Nanjing, China) and were passed through 40-μm nylon sterile strainer to obtain single-cell suspensions. To reduce red blood cells, GEXSCOPE Red Blood Cell Lysis Buffer (Singleron Biotechnologies) was added to the cell suspension. scRNA-seq libraries were constructed following the Singleron’s protocol using a GEXSCOPE Single-cell RNA kit (1110012). After quality checks, scRNA-seq libraries were pooled and sequenced on a NovaSeq 6000 instrument (Illumina) with 150 bp paired end reads. Sequencing outputs were demultiplexed to convert BCL files to FASTQ format using bcl2fastq. Sequencing data were processed using the CeleScope 1.1.7 pipeline (Singleron). To detect viral RNA, reads were aligned to a custom blended genome containing GRCh38.98 and SARS-CoV-2 (ASM985889v3). A viral transcript containing the entire SARS-CoV-2 genome was added to the GRCh38.98GTF files to enable detection of SARS-CoV-2.55 After STAR (2.6.1b) alignment and samtools (1.9) filtering,62,63 the filtered count matrix was separated by the cell type judgment result obtained by the CeleScope for further analysis. A total of 1551 cells with 1294 genes per cell as median were remained.
Normalization, clustering and cell type identification of scRNA-seq
Single-nucleus expression matrix by CeleScope was performed using the Seurat package (version3.2.1) for filtering, data normalization, dimensionality reduction, clustering, and gene differential expression analysis.64 Briefly, cells with high quality were selected with the following criteria: (1) cells with unique features < 200 or > 5000 were removed; (2) Cells with ≥ 20% mitochondrial counts were removed. For each cell, the counts were log normalized with the “NormalizeData” function. Two thousand variable genes were selected using the “FindVariableGenes” function. A KNN graph based on the euclidean distance was constructed in PCA space. The edge weights between any two cells were refined based on the shared overlap in their local neighborhoods. Cells were clustered using the “FindClusters” function at an appropriate resolution, and were visualized using a Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) algorithm with the “RunUMAP” function. Wilcoxon rank-sum test was performed for each cluster using the “findMarkers” function to look for differentially expressed genes. Cell type of each cluster was annotated by the known marker genes (Supplementary information, Table S3). Through the above analysis, we processed the scRNA-seq data of 1437 high-quality cells.
Statistical analysis
All statistical analyses were performed by using SPSS version 13.0 software (SPSS Inc.). Categorical variables were described as frequency rates and percentages, and continuous variables were described using mean, median, or interquartile range (IQR) values. Bivariate correlation analysis (Pearson r test) was used to examine the correlation of two variables in human specimens. All data met the assumptions of the tests.
References
Gerges, H. J. et al. SARS, MERS and COVID-19: clinical manifestations and organ-system complications: a mini review. Pathog. Dis. 78, ftaa033 (2020).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Grasselli, G. et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA 323, 1574–1581 (2020).
Wang, D. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323, 1061–1069 (2020).
Edler, C. et al. Dying with SARS-CoV-2 infection-an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int. J. Legal Med. 134, 1275–1284 (2020).
Schaller, T. et al. Postmortem examination of patients with COVID-19. JAMA 323, 2518–2520 (2020).
Liu, Q. et al. Pathological changes in the lungs and lymphatic organs of twelve COVID-19 autopsy cases. Natl. Sci. Rev. 7, 1868–1878 (2020).
Efe, I. E., Aydin, O. U., Alabulut, A., Celik, O. & Aydin, K. COVID-19-associated encephalitis mimicking glial tumor. World Neurosurg. 140, 46–48 (2020).
Bian, X. W. The COVID-19 Pathology Team Autopsy of COVID-19 patients in China. Natl. Sci. Rev. 7, 1414–1418 (2020).
Su, H. et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 98, 219–227 (2020).
Yu, P. et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp. Med. 3, 93–97 (2020).
Bao, L. et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583, 830–833 (2020).
Carsana, L. et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect. Dis. 20, 1135–1140 (2020).
Wang, C. F. et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine 57, 102833 (2020).
Meinhardt, J. et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 24, 168–175 (2021).
Puelles, V. G. et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 383, 590–592 (2020).
Menter, T. et al. Post-mortem examination of COVID19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings of lungs and other organs suggesting vascular dysfunction. Histopathology 77, 198–209 (2020).
Delorey, T. M. et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature https://doi.org/10.1038/s41586-021-03570-8 (2021).
Martines, R. B. et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg. Infect. Dis. 26, 2005–2015 (2020).
Xu, J. et al. Detection of severe acute respiratory syndrome coronavirus in the brain: potential role of the chemokine mig in pathogenesis. Clin. Infect. Dis. 41, 1089–1096 (2005).
Farcas, G. A. et al. Fatal severe acute respiratory syndrome is associated with multiorgan involvement by coronavirus. J. Infect. Dis. 191, 193–197 (2005).
Shieh, W. J. et al. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum. Pathol. 36, 303–309 (2005).
Nicholls, J. M. et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361, 1773–1778 (2003).
Franks, T. J. et al. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34, 743–748 (2003).
Gu, J. et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 202, 415–424 (2005).
Ding, Y. et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 203, 622–630 (2004).
Chau, T. N. et al. SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology 39, 302–310 (2004).
To, K. F. et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 202, 157–163 (2004).
Thille, A. W. et al. Chronology of histological lesions in acute respiratory distress syndrome with diffuse alveolar damage: a prospective cohort study of clinical autopsies. Lancet Respir. Med. 1, 395–401 (2013).
Zhou, F. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062 (2020).
Wang, K. et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 5, 283 (2020).
Radzikowska, U. et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 75, 2829–2845 (2020).
Tang, X. et al. Transferrin receptor is another receptor for SARS-CoV-2 entry. bioRxiv https://doi.org/10.1101/2020.10.23.350348 (2020).
Cantuti-Castelvetri, L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020).
Dorward, D. A. et al. Tissue-specific immunopathology in fatal COVID-19. Am. J. Respir. Crit. Care Med. 203, 192–201 (2021).
Shinu, P. et al. SARS CoV-2 organotropism associated pathogenic relationship of gut-brain axis and illness. Front Mol. Biosci. 7, 606779 (2020).
Ren, X. et al. COVID-19 immune features revealed by a large scale single cell transcriptome atlas. Cell 184, 1895–1913 (2021).
Trypsteen, W., Cleemput, J. V., Snippenberg, W. V., Gerlo, S. & Vandekerckhove, L. On the whereabouts of SARS-CoV-2 in the human body: a systematic review. PLoS Pathog. 16, e1009037 (2020).
Varga, Z. et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 395, 1417–1418 (2020).
Bonetti, P. O., Lerman, L. O. & Lerman, A. Endothelial dysfunction: a marker of atherosclerotic risk. Arterioscler. Thromb. Vasc. Biol. 23, 168–175 (2003).
Hadjadj, J. et al. Impaired type I interferon activity and exacerbated inflammatory responses in severe COVID-19 patients. Science 369, 718–724 (2020).
Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020).
Wang, W. et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844 (2020).
Zheng, S. et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ 369, m1443 (2020).
Li, H. et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet 395, 1517–1520 (2020).
Zhou, J. et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 209, 1331–1342 (2014).
Arvin, A. M. et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 584, 353–363 (2020).
Soin, A. S. et al. Tocilizumab plus standard care versus standard care in patients in India with moderate to severe COVID-19-associated cytokine release syndrome (COVINTOC): an open-label, multicentre, randomised, controlled, phase 3 trial. Lancet Respir. Med. 9, 511–521 (2021).
Chi, X. et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 (2020).
Wang, C. et al. Imaging mass cytometric analysis of postmortem tissues reveals dysregulated immune cell and cytokine responses in multiple organs of COVID-19 patients. Front Microbiol. 11, 600989 (2020).
Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 395, 1033–1034 (2020).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Merad, M. & Martin, J. C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 20, 355–362 (2020).
Cheung, C. Y. et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 79, 7819–7826 (2005).
Grant, R. A. et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 590, 635–641 (2021).
Wauters, E. et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 3, 272–290 (2021).
Wang, Y. et al. MBD2 serves as a viable target against pulmonary fibrosis by inhibiting macrophage M2 program. Sci. Adv. 7, eabb6075 (2021).
Tian, S. et al. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 15, 700–704 (2020).
Yao, X. H. et al. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 30, 541–543 (2020).
Segalés, J. et al. Detection of SARS-CoV-2 in a cat owned by a COVID-19-affected patient in Spain. Proc. Natl. Acad. Sci. USA 117, 24790–24793 (2020).
Liu, J. et al. Molecular detection of SARS-CoV-2 in formalin-fixed, paraffin-embedded specimens. JCI Insight 5, e139042 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Acknowledgements
We thank the patients and their families for their contributions to scientific research. We thank the staff from the Wuhan Jinyintan Hospital, Taikang Tongji Hospital, Zhongfaxincheng Hospital, and Huoshenshan Hospital for their support to this study. We thank Prof. Zhinan Chen, Drs. Yang Zhang and Xu Yang from the National Translational Science Center for Molecular Medicine & Department of Cell Biology, Fourth Military Medical University for providing Cercopithecus Vero E6 cells. We thank Prof. Chong Liu from the Department of Pathology and Pathophysiology, Zhejiang University School of Medicine for analyzing scRNA-seq data. This study was supported by the projects for COVID-19 from Chongqing Science and Technology Commission (cstc2020jscx-fyzxX0037 to X.-W.B.), Chongqing Health Committee (2020NCPZX01 to X.-W.B.), and the Third Military Medical University (2020XGBD08 to X.-W.B.).
Author information
Authors and Affiliations
Contributions
X.-W.B., Y.-F.P., and S.-Y.Z. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Y.S., Y.-F.P., Y.W., X.-D.L., and X.-W.B. contributed to writing of the manuscript. X.-H.Y., Z.-C.H., R.T., J.C., X.-D.Z., D.-P.J., X.-C.F., X.-Q.H., L.Z., H.Z., Z.-H.L., H.-B.W., Y.R., R.C., J.W., C.-L.Y., T.-S.L., W.-Q.L., C.-F.W., and Q.M. performed autopsy or collected the clinical information. T.L., Z.-C.H., W.-J.F., H.L., C.C., H.-R.Z., X.-Y.X., R.J., and Z.-X.Y. performed SARS-CoV-2 nucleic acid detection, tissue staining, electronic microscopy, or scRNA-seq. X.-W.B., X.-H.Y., Y.-F.P, P.-P.Z., H.-R.Z., and Y.S. conducted microscopic examination. D.-Y.Z. and Y.-Q.D. contributed to the revision of the report.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yao, XH., Luo, T., Shi, Y. et al. A cohort autopsy study defines COVID-19 systemic pathogenesis. Cell Res 31, 836–846 (2021). https://doi.org/10.1038/s41422-021-00523-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-021-00523-8
This article is cited by
-
Comprehensive analysis of immunogenic cell death-related gene and construction of prediction model based on WGCNA and multiple machine learning in severe COVID-19
Scientific Reports (2024)
-
Infection with SARS-CoV-2 can cause pancreatic impairment
Signal Transduction and Targeted Therapy (2024)
-
Systematic analyses of the factors influencing sperm quality in patients with SARS-CoV-2 infection
Scientific Reports (2024)
-
The unfolded protein response pathway as a possible link in the pathogenesis of COVID-19 and sepsis
Archives of Virology (2024)
-
A comprehensive SARS-CoV-2 and COVID-19 review, Part 2: host extracellular to systemic effects of SARS-CoV-2 infection
European Journal of Human Genetics (2024)