Abstract
The pituitary adenylate cyclase-activating polypeptide type I receptor (PAC1R) belongs to the secretin receptor family and is widely distributed in the central neural system and peripheral organs. Abnormal activation of the receptor mediates trigeminovascular activation and sensitization, which is highly related to migraine, making PAC1R a potential therapeutic target. Elucidation of PAC1R activation mechanism would benefit discovery of therapeutic drugs for neuronal disorders. PAC1R activity is governed by pituitary adenylate cyclase-activating polypeptide (PACAP), known as a major vasodilator neuropeptide, and maxadilan, a native peptide from the sand fly, which is also capable of activating the receptor with similar potency. These peptide ligands have divergent sequences yet initiate convergent PAC1R activity. It is of interest to understand the mechanism of PAC1R ligand recognition and receptor activity regulation through structural biology. Here we report two near-atomic resolution cryo-EM structures of PAC1R activated by PACAP38 or maxadilan, providing structural insights into two distinct ligand binding modes. The structures illustrate flexibility of the extracellular domain (ECD) for ligands with distinct conformations, where ECD accommodates ligands in different orientations while extracellular loop 1 (ECL1) protrudes to further anchor the ligand bound in the orthosteric site. By structure-guided molecular modeling and mutagenesis, we tested residues in the ligand-binding pockets and identified clusters of residues that are critical for receptor activity. The structures reported here for the first time elucidate the mechanism of specificity and flexibility of ligand recognition and binding for PAC1R, and provide insights toward the design of therapeutic molecules targeting PAC1R.
Similar content being viewed by others
Introduction
PACAP is a 38-amino acid C-terminally amidated polypeptide (PACAP38) that was discovered as a hypothalamic neuropeptide to potentially induce cAMP levels in anterior pituitary cells.1 The N-terminal 27 residues of PACAP38, highly conserved in almost all vertebrate species and responsible for the physiological activity of the peptide, undergo internal cleavage-amidation to generate the PACAP27 fragment. Because of the 68% sequence identity between PACAP27 and vasoactive intestinal polypeptide (VIP), PACAP is identified as a member of the glucagon/gastric inhibitory polypeptide (GIP)/secretin/VIP family—a hormone family consisting of evolutionarily related peptides that regulate the activity of class B G-protein coupled receptor (GPCR) family, also known as secretin receptor family.2
The receptors that recognize PACAP are characterized into three distinct subtypes based on their relative affinities to PACAP and VIP: the pituitary adenylate cyclase-activating polypeptide type I receptor (PAC1R) with two orders of magnitude higher affinity to PACAP than to VIP; the vasoactive intestinal polypeptide receptor 1 (VPACR1) and receptor 2 (VPACR2) with similar PACAP/VIP affinities.3
PACAP and its receptors are broadly expressed in the central nervous system (CNS) and in most peripheral organs, and have been found to exert a variety of functions including control of neurotransmitter release, vasodilation, bronchodilation, activation of intestinal motility, neuroprotection, immune modulation, and stimulation of cell proliferation and/or differentiation.4
As the major sensory and vasodilator neuropeptides, PACAP38 and VIP are involved in parasympathetic communication with the cranial vasculature. Abnormal activation and sensitization of the central trigeminovascular pain pathway mediate migraine and the release of these peptides.5 Intravenous infusion of PACAP38 but not VIP induces delayed migraine-like headaches, indicating that PAC1R is playing a major role in migraine6 and suggesting PACAP38-PAC1R as a potential therapeutic target for migraine treatment.7
Maxadilan, another natural PAC1R agonist, is a 61-amino acid polypeptide isolated from the salivary gland of the blood-feeding sand fly Lutzomia lingipalpis.8 Maxadilan is an immunomodulator and has been shown to facilitate the transmission and establishment of leishmaniasis. Despite the low sequence homology between maxadilan and PACAP38, they both potently activate PAC1R. The structural basis of receptor ligand recognition and activation mechanism remains unknown.
Recent development in cryo-electron microscopy (cryo-EM) has enabled the determination of full-length class B GPCR structures in complex with their peptide ligand and G protein complex.9,10,11,12,13 However, no structural information is available for PAC1R. Here we report the cryo-EM structures of Gs-protein coupled PAC1R in complex with PACAP38 and maxadilan, respectively, and uncover the underlying mechanism of convergent activity from distinct ligands on a class B GPCR. Structure-guided mutagenesis elucidates the key residues responsible for ligand binding and receptor activation. These results further strengthen our mechanistic understanding of PAC1R regulation and will benefit future rational design of therapeutic molecules for migraine.
Results
Structure determination
For cryo-EM structure determination purpose, we modified the human PAC1R with a short C-terminal truncation (439–468), seven mutations (T163L, T167A, T169L and T170L on TM1; T276A, T278L and C280F on TM4), replacement of the native signal peptide by that of haemagglutinin (HA), and addition of affinity tags (an N-terminal FLAG tag and a C-terminal 10× His tag) (Supplementary information, Fig. S1). These modifications did not alter receptor ligand binding and pharmacological properties (Supplementary information, Fig. S2). We generated by site-directed mutagenesis the dominant-negative Gαs construct, including mutations that reduce nucleotide affinity (S54N and G226A) and improve the dominant-negative effect (E268A, N271K, K274D, R280K, T284D, and I285T) to improve the complex stability.10
Human PAC1R, Gαs, Gβ1, and Gγ2 were co-expressed in High Five insect cells using baculovirus transfection to form the GPCR complex. Agonist peptide, PACAP38 or maxadilan, and Nanobody-35 (Nb35)9,10 were added during purification to enable a stable complex formation. The complex was solubilized in lauryl maltose neopentyl glycol (LMNG) and cholesteryl hemisuccinate, and then purified by nickel affinity and size exclusion chromatography to yield a monodisperse complex that contained all the components (Supplementary information, Fig. S2).
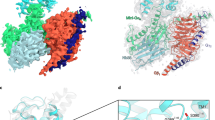
Single-particle cryo-EM analysis of the complexes in vitreous ice yielded a final map at a resolution of 3.5 Å for PACAP38-PAC1R-Gs (reconstructed from 82,970 particles) and that of 3.6 Å for maxadilan-PAC1R-Gs (reconstructed from 58,451 particles) (Fig. 1; Supplementary information, Figs. S3, 4, Table S1). The density for the seven transmembrane helices (TMs), the ligands, and G protein complex are unambiguously determined based on the well-traced α-helices and aromatic side chains. The extracellular domain (ECD) is incomplete due to flexibility, but we were able to utilize the previously solved crystal structure14 to place it according to the partial density (Supplementary information, Fig. S5).
a, b Cryo-EM density map (left), the structure model (middle) and local resolution distribution map (right) of PAC1R-Gs in complex with PACAP38 (a) and maxadilan (b). The structures of PAC1R (green in PACAP38-PAC1R-Gs structure, orange in maxadilan-PAC1R-Gs structure), PACAP38 (pink), maxadilan (purple), Gα (red), Gβ (blue), Gγ (yellow) and Nb35 (gray) are represented in cartoon.
Overall structures of the PAC1R-Gs complexes
PAC1R-Gs complexes, with PACAP38 or maxadilan bound at the extracellular side and Gs protein complex bound at the intracellular side of PAC1R, are in an active state (Fig. 1) with conformation reminiscent of the other class B GPCR-Gs complex structures.9,10,11,12,13 The overall reported resolution is 3.5 Å and 3.6 Å for the PACAP38-PAC1R-Gs and the maxadilan-PAC1R-Gs complexes, respectively (Fig. 1a, b). Although both structures represent G protein-bound active state of PAC1R, the ligands PACAP38 and maxadilan, with their dramatically different sequences and stereo structures, adopted disparate modes of binding to the ECD and the orthosteric site. To accommodate differences in ligand size, the TMs and extracellular loops (ECLs) that consist of the orthosteric site shift slightly with a root-mean-square deviation (r.m.s.d.) of 1.11 Å in the 7-TM region between the two structures.
PACAP38-PAC1R binding interface
The PAC1R construct used in the cryo-EM study retains ligand binding and pharmacological properties comparable to the wild-type (WT) receptor (Supplementary information, Fig. S2). The peptide forms an α-helix with its N-terminus inserted into the orthosteric site. Structure of the N-terminal 27 residues, conserved and responsible for receptor activation, are unambiguously resolved15 (Fig. 2a), while the C-terminal 11 residues are not visible in the map probably due to flexibility and omitted from the final reported structure.
a Slice view of the PACAP38-PAC1R-Gs structure and a schematic helical sequence for PACAP38 with unresolved residues in red. b Close view of the C-terminal part of PACAP38 showing detailed interactions. c Close views of the N-terminal part of PACAP38 from two angles with 180° rotation. The effective residues in mutagenesis assay are presented by spheres. d The PAC1R-PACAP38 interaction diagrams. PACAP38 (H1-Y13) is shown as sticks. Residues are represented as spheres and colored by interaction type. Interactions between the residues and the ligand atoms are drawn as dashed lines, colored by interaction type. The solvent accessible surface of an interacting residue and of an atom are represented by haloes around the residue and the atom. The diameter of the circle is proportional to the solvent accessible surface.
Outside the orthosteric site, PACAP38 mainly forms hydrophobic interactions with the receptor in the ECD region (residues 53–88) that is previously reported to be critical for ligand binding affinity16: K15P (PACAP38) and V19P hydrophobically interact with I83ECD and F84ECD of the β3–β4 loop; V26P forms hydrophobic alkyl interaction with I61ECD of the β1–β2 loop (Fig. 2b).
Within the orthosteric site, PACAP38 interacts with PAC1R through TM1, TM2 and ECL1 on one side and TM3 and ECL2 on the other side. On TM1, Y1501.36 forms hydrogen bond with S9P and hydrophobic interaction with Y10p and Y13P. D1451.31 forms hydrogen bond with Y13P. On TM2, PAC1R L2102.71 forms hydrophobic interactions with Y10P and establishes hydrogen bond with R14 via its main chain. ECL1 closely attaches to PACAP38, but under current resolution we cannot resolve specific residue interactions (Fig. 2c). On the other side, D298ECL2 forms hydrogen bond with S11p. M299ECL2 interacts with R12P and K15P hydrophobically. In addition, W3065.39 may form hydrophobic interactions with H1P and I5P. At the bottom of the orthosteric site, Y1611.47 and R1992.60 form hydrogen bonds with D3P (Fig. 2c, d; Supplementary information, Table S2).
Maxadilan-PAC1R binding interface
Maxadilan is a similar ligand compared to PACAP38 in terms of ligand affinity and receptor activation, however, the structure is very different (Supplementary information, Fig. S2). Maxadilan forms the N- and C-terminal helices that are linked by a loop, deletion of which converts the peptide into an antagonist (Fig. 3a).17 In model building, we defined the residues of the helices based on the disulfide bond between C14M (maxadilan) and C51M,18 as well as the side chain densities of aromatic residues (Fig. 3a). In the maxadilan-PAC1R-Gs structure, PAC1R ECD adopts a dramatically different conformation from that of the PACAP38-PAC1R-Gs structure, but intriguingly utilize the same interface for maxadilan binding. Some detailed interactions are not well defined because of the incomplete ECD map; the map for aromatic residues such as F7M and F59M, and Y130ECD, F131ECD and F136ECD on the α3 helix is of great quality and reveals hydrophobic interaction network among these residues (Fig. 3b).
a Slice view of the maxadilan-PAC1R-Gs structure and a schematic helix-loop-helix sequence for maxadilan. b Close view of maxadilan from outside of the binding pocket showing detailed interactions. c Close views of the insides of the orthosteric site from both sides with 180° rotation. The effective residues in mutagenesis assay are presented by spheres. d The PAC1R-Maxadilan interaction diagrams. Maxadilan (H19-L42) is shown as sticks. Residues are represented as spheres and colored by interaction type. Interactions between the residues and the ligand atoms are drawn as dashed lines, colored by interaction type. The solvent accessible surface of an interacting residue and of an atom are represented by haloes around the residue and the atom. The diameter of the circle is proportional to the solvent accessible surface.
Within the orthosteric site, maxadilan, bearing a much larger size and a distinct shape in contrast to PACAP38, establishes different interactions with the receptor. On one side, PAC1R closely interacts with the C-terminal helix of maxadilan through TM1 and ECL1. On TM1, D1471.33, Y1501.36 and K1541.40 form hydrogen bonds with S21M, Q25M and S27M, respectively. Y1501.36 and V1531.39 interact with V28M through hydrophobic interactions. On ECL1, Q214ECL1 backbone forms hydrogen bond with H19M whose side chain hydrophobically interacts with F220ECL1 (Fig. 3c). On the other side of maxadilan, the loop wedges into the orthosteric site to interact with ECL2, TM6 and TM7. On ECL2, N300ECL2 interacts with S36M and M37M (backbone) hydrophilically, and D301ECL2 forms hydrogen bonds with T39M. On TM6, the backbones of F3696.56 and A3706.57 stabilize F34M through amide-Pi stacked interaction. On TM7, R379ECL3 forms hydrogen bonds with Q25M and L3867.43 hydrophobically interacts with A32M (Fig. 3c, d; Supplementary information, Table S2).
PAC1R activation and G protein engagement
Peptide ligands associate with the large ECD of class B GPCRs and interact with the transmembrane bundles to activate the receptors. The interaction induces reorganization of the buried polar residues in the TM bundle, which is believed to reflect the mode of receptor activation.19,20,21 Three layers of highly conserved polar networks governed this process: (i) on the top is the central polar network formed by residues R2.60, N3.43, H6.52 and Q7.49,19,21 (ii) in the middle is the HETX polar networks formed by H2.50, E3.50, T6.42, Y7.57, (iii) at the bottom is the TM2-6-7-8 network formed by R2.46, R6.37, N8.47, E8.49 (Supplementary information, Fig. S6).20 Similar to other class B GPCRs, dissociation of the central hydrogen bond network upon peptide binding induces destabilization of TM6 around the P6.47XXG6.50 motif, which enables further rearrangement in the lower polar networks to facilitate receptor activation and G protein engagement. Upon dissociation of the HETX polar network, the side chain of Y4007.57 in both PAC1R structures shifts away and T3556.42 further moves away along with the bended TM6 (Supplementary information, Fig. S6). Mutations at T3556.42 to destabilize the HETX network and at the P6.47XXG6.50 motif to stabilize the active conformation both result in constitutive activation of PAC1R (data not shown).
Upon peptide ligand-induced activation, PAC1R engages Gs heterotrimeric protein to the transmembrane bundle on the cytoplasmic side with a cavity, formed by outward bending of TM6, to interact with the Gαs Ras-like domain. This extensive interface consists of both hydrophobic and electrostatic interactions in a similar pattern as reported in other Gαs-class B GPCR complexes.9,10 These interactions are summarized in Supplementary information, Fig. S7 and Table S3. We report more interactions from the PACAP38-PAC1R-Gs mainly due to the better map quality of this structure. A key difference in the activation of class B GPCRs comes from the additional hydrophilic interactions formed between helix 8 (H8) and Gβ protein, compared to class A GPCRs.11 In PAC1R, we observed this conserved interaction between R4138.56, K180ICL1 and H311, D312 on Gβ, respectively (Supplementary information, Fig. S7 and Table S3). Like other class B GPCRs in active conformation, H8 interacts with detergent micelle with bulky aromatic residues on the membrane-proximal face (Supplementary information, Fig. S7).9,10
PAC1R ligand binding specificity and flexibility
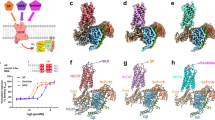
To identify critical residues responsible for ligand-induced receptor activation, we conducted virtual alanine scanning on all the residues within the ligand-binding pockets and evaluated the destabilizing effects through mutation energy analysis (Supplementary information, Fig. S8). We then selected the most effective residues on the mutation energy ordering (> 0.5 kcal/mol) and key interacting residues derived from structural analysis, designed single point alanine substitutions and tested their effects on PAC1R activation experimentally (Supplementary information, Table S4).
Y130ECDA and F131ECDA showed significant decreases in EC50 of maxadilan- but not PACAP38-induced receptor activity in the functional cAMP assay, and both mutations, particularly F131A, failed to compete for I125-PACAP27 binding in the competition binding assay, indicating that the hydrophobic interactions are crucial for binding of maxadilan to induce receptor activation (Fig. 3b; Supplementary information, Table S4 and Fig. S9). Y1571.43A showed decrease of EC50 of maxadilan-induced receptor activity, consistent with the maxadilan-PAC1R-Gs structure where Y1571.43 is close to the loop region of maxadilan (Fig. 3c). Intriguingly, the alanine substitution of R1992.60 specifically affected PACAP38-induced receptor activity and absolutely lost binding affinity to I125-PACAP27 (Supplementary information, Table S4 and Fig. S9). It is consistent with the structural observation that PACAP38 binds much deeper than maxadilan and establishes additional interaction through hydrogen bond with D3P (Fig. 2c; Supplementary information, Table S2). Y1611.47, with side chain pointing to the same position as R1992.60, showed similar preference of PACAP38 function (Fig. 2c; Supplementary information, Table S2). Both indicate that PACAP38 requires additional interactions on its N-terminus for ligand binding and function. In GLP1 receptor, K1972.67 and D1982.68 hold the aromatic moiety of Y1481.43 in an optimal position for ligand binding and receptor activity.22 Similarly, alanine substitutions of the corresponding residues K2062.67 and D2072.68 in PAC1R lost receptor activity with both ligands (Supplementary information, Table S4). In GLP1 receptor, mutation of W3065.36 to alanine results in reduced potency by > 200-fold.22,23,24 Similarly, in PAC1R W3065.36A led to significant decrease of activities to both ligands (Supplementary information, Table S4).
The density map of M299ECL2 and D301ECL2 did not allow unambiguous fitting of side chain and interpreted direct interactions with maxadilan, but alanine substitution of either M299 or D301 specifically disturbed maxadilan-induced receptor activation. It might alter the conformation of ECL2 that is an important region for peptide ligand recognition.23,24 L3827.39A and E3857.42A significantly and specifically decreased the potency of maxadilan-induced receptor activation, but no structural relevance was observed from the structure (Supplementary information, Table S2). They might alter the binding pocket, thus specifically disturbing the mode by which maxadilan induced receptor activation.
PAC1R ECD displays dramatic conformational flexibility to accommodate two stereo distinct ligands, PACAP38 and maxadilan. Both peptides utilize the same ECD interface, consisting of the β1–β2 loop, the β3–β4 loop, and the α2 helix, for binding, resulting in different PAC1R ECD orientations (Fig. 4a). In both ligand binding modes, the TM1-ECD loop half embraces the peptide blocking it from escaping once the ligand inserts into the orthosteric site. These structural features demonstrate a ligand recognition and binding mechanism where ECD fits the divergent peptide orientation with flexible conformation and holds the peptide in the orthosteric site with steric hindrance (Fig. 4a). In line with the size and binding orientation of ligands, the ligand-binding pocket consisting of the upper parts of TMs and ECLs was also slightly reorganized. TM6, ECL3, and TM7 slightly shift outward in the PACAP38 structure, whereas TM1, ECL1 and TM2 in the maxadilan structure outward shift to fit for the extended conformation of maxadilan N-terminal helix (Fig. 4a). Maxadilan wedged the ligand-binding pocket more open by ~6° (Fig. 4b), demonstrating the flexibility for PAC1R ligand recognition.
a Superposition of PACAP38-PAC1R-Gs (green cartoon) and maxadilan-PAC1R-Gs (orange cartoon) structures. Close views from outside of the cell membrane (red squares) showing the PACAP38 and maxadilan binding in the orthosteric site. Close view (blue square) showing that the TM1 and TM2 shift outward in maxadilan binding mode. b Slice side views of the receptor, in parallel to the cell membrane, showing the binding pocket for PACAP38 (pink) and maxadilan (purple). The angels of the ‘cleft’ between TM2–5 and TM1, 6–7 are shown by dash lines.
Discussion
PAC1R recognizes and binds the ligand as a ‘glove’, with the ‘palm’ (ECD) and the ‘thumb’ (ECL1) holding the ligand in the orthosteric site through hydrophilic and hydrophobic interactions (Fig. 5). ECD is flexible to fit for different orientations of ligand and connects to TM1 with a long loop which embraces the ligand and keeps it from escaping. ECL1 interacting with the ligand on the other side further stabilizes ligand binding (Fig. 5). The holding conformation of ECD and ECL1 enables PACAP38 or maxadilan to insert into the orthosteric site and form similar interactions for receptor activation. This ‘glove’ ligand binding mechanism is reminiscent of the two-domain binding hypothesis proposed for PAC1R,25 where the ECD creates a locally high concentration of ligand through high-affinity interactions with the ECD binding part of the ligand and further facilitates the orthosteric binding part to insert into the orthosteric site.
Close views showing that ECD and ECL1 in different orientations ‘holding’ the peptides are highlighted in blue circles. The helices in PACAP38 and maxadilan are shown as pink and purple rods. The conformations of ECL1 in two structures are shown in cartoon. The relative positions of ECD and ECL1 are presented in circles and lines, revealing the flexibilities for different ligand binding. The hydrophilic interaction for PACAP38 is represented by triangle and the hydrophobic interaction for maxadilan is represented by hexagon.
Although PACAP38 and maxadilan activate the PAC1R in similar potencies and induce similar conformational changes for G protein engagement, there are subtle differences in ligand binding and receptor activation reflected by the structures and functional assays. PACAP38 requires R1992.60 at the bottom of the pocket for binding to the pocket and could use this interaction to affect the central polar network for receptor activation. As for maxadilan, the loop does not insert into the pocket as deep as PACAP38 to reach R1992.60, whereas the interface between the loop and TM7 is likely an important region for receptor activation as confirmed by mutagenesis of L3827.39A and E3857.42A. These structural insights explain the previously reported results that maxadilan, with Q25–Q41 deleted, was an antagonist. This deleted form of maxadilan will not be able to make interactions with critical residues on TM7 or insert into the orthosteric site, hence fail to activate the downstream elaborate polar networks.17 In contrast to PACAP38, maxadilan is unlikely to rely on the residues within the pocket for ligand binding but relies on binding to the ECD through hydrophobic interaction with F131ECD. These observed subtle differences in ligand binding will be useful for fine-tuning the affinity and activity of PACAP38 and maxadilan for therapeutic purposes.
We compared PACAP38-PAC1R-Gs structure to previously solved structures of class B GPCRs bound with single-helix peptide ligand to try to understand the complex sequence-structure relationship. Sequence alignment indicates that GLP1 shows some similarity to PACAP38, whereas PTH shows barely any sequence resemblance to PACAP38 (Supplementary information, Fig. S10a). However, structure wise, PTH-PTH1R-Gs has a more similar peptide binding orientation and the ECD and ECL1 conformations are almost identical to PACAP38-PAC1R-Gs, whereas in the GLP1-GLP1R-Gs structure, GLP1 peptide slightly swings away and the ECD and ECL1 conformations are dramatically different (Supplementary information, Fig. S10b). CGRP, with unstructured loops in both N- and C- terminal regions, is totally different from PACAP38-PAC1R-Gs by both sequence and structural alignments (Supplementary information, Fig. S10b).
Conclusion
Delineating the properties of ligand binding specificity and flexibility of PAC1R may guide rational design of novel molecules with improved specificity and desired therapeutic effects. We report cryo-EM structures of PACAP38-PAC1R-Gs and maxadilan-PAC1R-Gs complexes, revealing two distinct PAC1R ligand binding modes. These structures reveal the conformational plasticity of ECD to recognize distinct ligands, and the specificity and flexibility of PAC1R ligand binding site to accommodate different ligands towards inducing convergent receptor activity in different modes. By molecular modeling, structure-guided mutagenesis and functional assay, we characterized and identified critical residues for ligand-induced receptor activation. These results provide the structural basis for PAC1R ligand recognition and bring insights into the receptor activation mechanism.
Materials and methods
Constructs
The human PAC1R was modified with an N-terminal deletion (89–109) that is a PAC1R splice variant with similar affinities to PACAP.16 A short C-terminal truncation (439–468) and seven mutations, T163L, T167A, T169L, T170L, T276A, T278L, C280F were introduced to increase protein yield and stability in micelle. The construct was cloned into both mammalian and insect cell expression vectors with replacement of the native signal peptide by that of haemagglutinin (HA) and addition of affinity tags (an N-terminal FLAG tag epitope and a C-terminal 10× His tag). These modifications did not alter receptor pharmacology (Supplementary information, Fig. S2). A dominant-negative Gαs (DNGαs) construct was generated by site-directed mutagenesis to incorporate mutations that reduce nucleotide affinity (S54N and G226A) and improve the dominant-negative effect (E268A, N271K, K274D, R280K, T284D, and I285T). The DNGαs was reported to have enhanced interaction between Gα and Gβγ subunits and improved overall trimeric complex stability.10 The 8× His-tagged human Gβ1 and human Gγ2 were cloned into pFastBac-dual vector.
Expression
Human PAC1R, human DNGαs, and 8× His-tagged human Gβ1 and human Gγ2 were expressed in High Five insect cells using baculovirus. Cell cultures were grown in ESF921 serum-free medium to a density of 2.5 × 106 viable cells/mL and then infected with three separate baculoviruses at a ratio of 1:1:1 for PAC1R, DNGαs, and Gβ1γ2. The culture was collected by centrifugation 48 h after infection and cell pellets were stored at –80 °C. Nanobody-35 (Nb35) was expressed in the periplasm of Escherichia coli strain WK6, extracted, and purified by nickel affinity chromatography according to previously described methods.26
WT PAC1R and mutations used in the functional assay were cloned into pJIF1.1 for BacMam virus generation.
Complex purification
The cell pellet was thawed in 20 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM MgCl2 supplemented with cOmplete Protein Inhibitor Cocktail tablets (Roche). Complex formation was initiated by addition of 10 μM PACAP38, Apyrase (25 mU/mL, NEB) and Nb35-His (10 μg/mL), the suspension was incubated for 1 h at 4 °C. Complexes from membranes were solubilized by 1% (w/v) lauryl maltose neopentyl glycol (L-MNG, Anatrace) complemented with 2 mM cholesteryl hemisuccinates (CHS, Anatrace) for 2 h at 4 °C. Insoluble material was removed by centrifugation at 40,000 rpm for 30 min and the solubilized complex was immobilized by batch binding to Ni-NTA resin (Qiagen). The resin was packed into a disposable plastic column (Bio-Rad) and washed with 20 column volumes of 20 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 0.01% (w/v) L-MNG, 20 μM CHS, and 50 mM Imidazole, and eluted with 4 column volumes of 20 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 0.01% (w/v) L-MNG, 20 μM CHS, and 300 mM Imidazole. The PAC1R-DNGs-Nb35 complex was then concentrated using an Amicon Ultra Centrifugal Filter (MWCO 100 kDa) before being subjected to size exclusion chromatography on a Superose 6 Increase 10/300 column (GE Healthcare) pre-equilibrated with 20 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 0.01% (w/v) L-MNG and 20 μM CHS to separate complex from contaminants. Eluted fractions consisting of monomeric receptor and G-protein complex were pooled and concentrated for electron microscopy experiments. The final yield of the purified complex was ~1 mg/L from insect cell culture.
Samples collected from each purification step were analyzed by SDS-PAGE. Precast gradient TGX gels (Bio-Rad) were used and stained by SimplyBlue (Invitrogen).
Cryo-EM sample preparation and data collection
EM grids (Quantifoil, 300 mesh golden R1.2/1.3) were glow discharged for 20 s using Harrick plasma cleaner (Harrick). Vitrified specimen was prepared by applying 3.5 μL of 5 mg/mL protein complex solution on the grid in the Vitrobot chamber (FEI Vitrobot Mark IV) with blotting time of 3 s. The chamber of Vitrobot was set to 100% humidity at 18 °C. Cryo-EM data were collected on a Titan Krios electron microscope operated at 300 kV equipped with a Gatan K2 Summit direct electron detection camera (Gatan) using AutoEMation.27 Micrographs were recorded in super-resolution mode at a nominal magnification of 105,000×, resulting in a physical pixel size of 0.5455 Å per pixel. Defocus values varied from –1.5 μm to –2.5 μm. The dose rate was 8.0 electron per pixel per second. Exposures of 5.6 s were dose-fractionated into 32 sub-frames, leading to a total accumulated dose of 50 electrons per Å2 on the specimen.
Image processing and 3D reconstruction
The raw super-resolution dose-fractionated image stacks were 2× Fourier binned, aligned, dose-weighted and summed using MotionCorr2.28 Contrast transfer function (CTF) parameters were estimated using CTFFIND4.29 Bad micrographs were removed manually based on the CTF parameters. The following processing steps were performed in RELION.30 For all the datasets, manually picked sets of particles were subjected to 2D classification. These generated templates for reference-based particle picking, respectively. Several rounds of reference-free 2D classification were performed to remove contaminants and bad particles from the automatically picked particle datasets. Initial models were generated using the “3D initial model” panel in RELION-3.0. After several rounds of reference-based 3D classification, the most homogeneous particles were selected for the final 3D auto-refinement. Local resolution distribution was estimated using “blocres” command in BSOFT software package.31 More details related to data processing are summarized in Supplementary information, Figs. S3, 4, Table S1.
Model building and refinement
An initial model was generated by homology modeling using the GLP1-GLP1R-Gs cryo-EM structure (PDB: 5VAI), and the ECD model also refers to the isolated GLP1R ECD structure (PDB: 3IOL) and PAC1R ECD structure (PDB: 3N94). Manual building and adjustment were performed in Coot.32 DNGα, Gβ, Gγ and NB35 models were taken from ExP5-GLP1R-Gs structure (PDB: 6B3J). The ECD of maxadilan-PAC1R-Gs (PDB: 3N94) was rigid-body docked into the model with manual adjustment owing to limited density. The final models were subjected to real space refinement and minimization using PHENIX.33 Model validation was performed using MolProbity.34
Molecular modeling
The structure preparation and virtual mutation calculation were performed with Discovery Studio (Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016). The PACAP38-PAC1R-Gs and the maxadilan-PAC1R-Gs complex structures were protonated at pH 7.4 with “Prepare Protein” protocol. Then the interactions between PAC1R and peptide ligands were analyzed with “Analyze Protein Interface” protocol. The identified interface residues were virtually mutated to Ala and for every single mutant, the differences in the free energy of binding between the WT and mutated structures are calculated with “Calculate Mutation Energy (Binding)” protocol. The “Effect of Mutation” is defined by default setting: Stabilizing, mutation energy is less than –0.5 kcal/mol; Neutral, mutation energy is between –0.5 and 0.5 kcal/mol; Destabilizing, mutation energy is greater than 0.5 kcal/mol.
The receptor-ligand interaction diagrams were generated for part of the ligands (PACAP38: H1-Y13; maxadilan: H19-L42) and PAC1R residues at the binding interface, with “Draw 2D Ligand Interaction Diagram” tool in Discovery Studio.
Transient expression by BacMam virus
CHO-K1 cells were cultured in DMEM supplemented with 10% FBS and infected with BacMam viruses with a MOI of 100. 24 h after infection, cells were collected for membrane preparation or functional assay.
Functional cAMP assay
Activation of WT PAC1R and mutants was measured based on intracellular cAMP levels using Lance Ultra cAMP kit (PerkinElmer) according to the manufacturer’s protocol. Briefly, in a well of 384-well plate, 1000 cells in 5 μL assay buffer (HBSS buffer pH 7.4 with 5 mM HEPES, 0.1% BSA and 0.5 mM IBMX) were mixed with 5 μL of different concentrations of PACAP38 or Maxadilan in assay buffer, incubated for 30 min at 37 °C. Then 5 µL of 4× Eu-cAMP and 5 µL of 4× Ulight-Anti-cAMP working solutions were added to each well and incubated at room temperature for 60 min before reading with Envision (PerkinElmer). All signals (ratio of 665 nm/615 nm) were fit with a sigmoidal dose-response model using GraphPad Prism 7.
Membrane preparation and radioligand binding assay
CHO-K1 cells were infected with BacMam virus using method mentioned above. Cells were then pelleted and membrane was generated following the protocol by Ban et al.35 In radioligand saturation binding assay, membrane of WT PAC1R or PAC1R mutants was incubated with a dose concentration of 125I-PACAP27, starting from 1 nM, in binding buffer (50 mM HEPES, pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 0.2% BSA). In radioligand competition binding assay, membrane of WT PAC1R or PAC1R mutants was incubated with a dose concentration of PACAP38 or Maxadilan in binding buffer containing 0.2 nM 125I-PACAP27. Binding reaction system was incubated at 30 °C for 2 h with gentle shaking, then filtered in UniFilter GF/B filtration Plate (PEI Coated, PerkinElmer), and washed three times immediately with ice-cold washing buffer (50 mM HEPES, pH 7.4, 500 mM NaCl, 0.1% BSA) using FilterMate™ Universal Harvester (PerkinElmer). After the plates were dried at 37 °C for 2 h, scintillation cocktail was added to each well, and radioactivity was counted in MicroBeta Trilux (PerkinElmer). Non-specific binding was determined in the presence of 100 nM PACAP38. All data were fit with one-site binding model using GraphPad Prism 7.
Change history
23 March 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Miyata, A. et al. Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem. Biophys. Res. Commun. 164, 567–574 (1989).
Segre, G. V. & Goldring, S. R. Receptors for secretin, calcitonin, parathyroid hormone (PTH)/PTH-related peptide, vasoactive intestinal peptide, glucagonlike peptide 1, growth hormone-releasing hormone, and glucagon belong to a newly discovered G-protein-linked receptor family. Trends Endocrinol. Metab. 4, 309–314 (1993).
Harmar, A. J. et al. Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide: IUPHAR review 1. Br. J. Pharmacol. 166, 4–17 (2012).
Vaudry, D. et al. Pituitary adenylate cyclase-activating polypeptide and its receptors: 20 years after the discovery. Pharmacol. Rev. 61, 283–357 (2009).
Waschek, J. A., Baca, S. M. & Akerman, S. PACAP and migraine headache: immunomodulation of neural circuits in autonomic ganglia and brain parenchyma. J. Headache Pain 19, 23 (2018).
Akerman, S. & Goadsby, P. J. Neuronal PAC1 receptors mediate delayed activation and sensitization of trigeminocervical neurons: relevance to migraine. Sci. Transl. Med. 7, 308ra157 (2015).
Rubio-Beltran, E. et al. PACAP38 and PAC1 receptor blockade: a new target for headache? J. Headache Pain 19, 64 (2018).
Lerner, E. A., Ribeiro, J. M., Nelson, R. J. & Lerner, M. R. Isolation of maxadilan, a potent vasodilatory peptide from the salivary glands of the sand fly Lutzomyia longipalpis. J. Biol. Chem. 266, 11234–11236 (1991).
Zhang, Y. et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253 (2017).
Liang, Y. L. et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555, 121–125 (2018).
Liang, Y. L. et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546, 118–123 (2017).
Liang, Y. L. et al. Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature 561, 492–497 (2018).
Zhao, L. H. et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science 364, 148–153 (2019).
Kumar, S., Pioszak, A., Zhang, C., Swaminathan, K. & Xu, H. E. Crystal structure of the PAC1R extracellular domain unifies a consensus fold for hormone recognition by class B G-protein coupled receptors. PLoS ONE 6, e19682 (2011).
Waschek, J. A. VIP and PACAP: neuropeptide modulators of CNS inflammation, injury, and repair. Br. J. Pharmacol. 169, 512–523 (2013).
Dautzenberg, F. M., Mevenkamp, G., Wille, S. & Hauger, R. L. N-terminal splice variants of the type I PACAP receptor: isolation, characterization and ligand binding/selectivity determinants. J. Neuroendocrinol. 11, 941–949 (1999).
Uchida, D. et al. Maxadilan is a specific agonist and its deleted peptide (M65) is a specific antagonist for PACAP type 1 receptor. Ann. N. Y. Acad. Sci. 865, 253–258 (1998).
Lerner, E. A., Iuga, A. O. & Reddy, V. B. Maxadilan, a PAC1 receptor agonist from sand flies. Peptides 28, 1651–1654 (2007).
Wootten, D. et al. A hydrogen-bonded polar network in the core of the glucagon-like peptide-1 receptor is a fulcrum for biased agonism: lessons from class B crystal structures. Mol. Pharm. 89, 335–347 (2016).
Wootten, D. et al. Key interactions by conserved polar amino acids located at the transmembrane helical boundaries in Class B GPCRs modulate activation, effector specificity and biased signalling in the glucagon-like peptide-1 receptor. Biochem. Pharmacol. 118, 68–87 (2016).
Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl. Acad. Sci. USA 110, 5211–5216 (2013).
Jazayeri, A. et al. Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546, 254–258 (2017).
Graaf, C. et al. Glucagon-like peptide-1 and its class B G protein-coupled receptors: a long march to therapeutic successes. Pharmcol. Rev. 68, 954–1013 (2016).
Dods, R. L. & Donnelly, D. The peptide agonist-binding site of the glucagon-like peptide-1 (GLP-1) receptor based on site-directed mutagenesis and knowledge-based modelling. Biosci. Rep. 36, e00285 (2015).
Hoare, S. R. Allosteric modulators of class B G-protein-coupled receptors. Curr. Neuropharmacol. 5, 168–179 (2007).
Rasmussen, S. G. et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69–80 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Ban, T. et al. GPCR structure and function relationship: identification of a biased apelin receptor mutant. Biochem. J. 475, 3813–3826 (2018).
Acknowledgements
We thank Jianlin Lei and Xiaomin Li at the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for help in cryo-EM data collection. We acknowledge the computational facility support on the cluster of Bio-Computing Platform at the Tsinghua University Branch of China National Center for Protein Sciences (Beijing).
Author information
Authors and Affiliations
Contributions
Y.M. and H.W. supervised the project. X.S. and X.C. developed the expression and purification strategy, and insect cell expression, and R.R. designed and optimized the constructs; X.S. performed protein purification and complex reconstitution, and prepared samples for cryo-EM; D.Z. performed complex purification; J.W. and X.S. performed imaging and data collection, electron microscopy data processing and analysis; C.L. and X.L. performed computational modeling and designed the mutations for functional assay; Y. Sun and Y. Song performed functional assay. Y.D. purified the NB35 nanobody; E.H. synthesized the ligand peptides; X.S. performed model building, refinement and validation. L.H., W.Z., and C.X. guided the project; X.S. wrote the manuscript with inputs from all the authors. X.H. contributed to the manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, J., Song, X., Zhang, D. et al. Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism. Cell Res 30, 436–445 (2020). https://doi.org/10.1038/s41422-020-0280-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-020-0280-2
This article is cited by
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Understanding VPAC receptor family peptide binding and selectivity
Nature Communications (2022)
-
A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2
Nature Communications (2022)
-
GPCR Intracellular Loop Regulation of Beta-Arrestin-Mediated Endosomal Signaling Dynamics
Journal of Molecular Neuroscience (2022)
-
A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor
Cell Research (2020)