Abstract
RNA interference (RNAi) has been thought to be a gene-silencing pathway present in most eukaryotic cells to safeguard the genome against retrotransposition. Small interfering RNAs (siRNAs) have also become a powerful tool for studying gene functions. Given the endosymbiotic hypothesis that mitochondria originated from prokaryotes, mitochondria have been generally assumed to lack active RNAi; however, certain bacteria have Argonaute homologs and various reports suggest the presence of specific microRNAs and nuclear genome (nDNA)-encoded Ago2 in the mitochondria. Here we report that transfected siRNAs are not only able to enter the matrix of mitochondria, but also function there to specifically silence targeted mitochondrial transcripts. The mitoRNAi effect is readily detectable at the mRNA level, but only recordable on relatively unstable proteins, such as the mtDNA-encoded complex IV subunits. We also apply mitoRNAi to directly determine the postulated crosstalk between individual respiratory chain complexes, and our result suggests that the controversial observations previously made in patient-derived cells might result from differential adaptation in different cell lines. Our findings bring a new tool to study mitochondrial biology.
Similar content being viewed by others
Introduction
Small RNAs, including small interference RNA (siRNA), microRNA (miRNA), and piRNA, play critical roles in regulating gene expression in eukaryotic cells. The RNA interference (RNAi) pathway has been well elucidated in most eukaryotic cells.1,2,3,4,5,6 Even lower eukaryotes, such as various yeast, have active RNAi, although certain yeast species, such as budding yeast, have lost such capability, which can be restored by re-expressing specific enzymes in the RNAi pathway.1 RNAi has been leveraged to study gene functions in diverse biological contexts. Interestingly, however, it has been unclear, even to date, whether an RNAi-like mechanism could operate in the mitochondria, which are thought to originate from parasitic bacteria.7 The conundrum is that certain bacteria are known to have homologs of Argonaute, a key enzyme in the RNA-induced silencing complex (RISC) responsible for targeted RNA degradation in higher eukaryotic cells,8 and that we and others have demonstrated that specific miRNAs are not only detectable, but also functional in the mitochondria.9,10 As miRNAs and siRNAs basically utilize the same machineries for their processing and function, the question is whether an RNAi-like mechanism could also function in the mitochondria, and if so, why it has never been reported in literature.
Mitochondria are endosymbiotic organelles in eukaryotic cells, which function as a powerhouse to generate ATP through oxidative phosphorylation (OXPHOS). Interestingly, unlike all living organisms whose genome size is largely proportional to their organismal complexity, the mitochondrial genome becomes minimized during evolution. The human mitochondrial genome, for example, is only ~16 kb in length and encodes 2 essential rRNAs and a minimal set (22) of tRNAs for translation as well as 13 polypeptides, all of which are membrane proteins as part of the respiratory chain complexes.11 Essentially all biochemical activities in mitochondria require nuclear genome (nDNA)-encoded proteins.12 While many nDNA-encoded proteins carry specific signal sequences to enter the mitochondria, at least 30% of them lack such signals,13 and whether certain RNAs can enter mitochondria has been an open, highly debatable question.14,15 Many lower eukaryotes clearly need import of tRNAs for mitochondrial translation, as some mtDNAs do not even encode any tRNA16 while others miss some key ones.17 Given the increasing evidence against mitochondrial RNA import, exemplified by the RNA component of RNase P and 5S rRNA,18,19 the field has been left with a general impression that RNA import into mitochondria has been permanently lost in mammalian cells.20 The lack of reliable functional readouts for potential RNA import has basically dampened the field towards a resolution to this fundamental question, not to mention the lack of an effective tool for direct perturbation of mtDNA-encoded genes.
In this study, we sought to systematically investigate whether siRNAs and small hairpin RNAs (shRNAs) are able to target mtDNA-encoded transcripts. Interestingly, we found that while randomly designed siRNAs do not always show anticipated effects, those designed following mapped argonaute 2 (Ago2)-binding peaks are all able to specifically target the intended mtDNA-encoded transcripts in an Ago2-dependent manner. We further show that most mitochondrial proteins are remarkably stable, thus preventing the detection of the anticipated RNAi effects at the protein levels, which might explain the lack of report on active mitoRNAi to date. We illustrate the use of this newly established siRNA tool to address some key questions in mitochondrial biology, which has been relying on the limited resource of patient-derived cells. Active mitoRNAi may also serve as an activity-based platform for future dissection of RNA import mechanism in the mitochondria.
Results
Localization of exogenous small RNAs in mitochondrial matrix
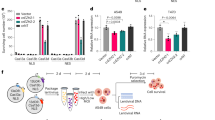
Multiple groups have reported the detection of endogenous small RNAs in the mitochondria.9,21 To determine whether exogenous small RNAs are able to enter the mitochondria, we transfected a Cy3-labeled siRNA of 21 nt into C2C12 cells. We purified mitochondria from transfected cells and showed the enrichment of mitochondrial matrix proteins (i.e., HSP60 and MRPL44). Trypsin treatment removed specific outer membrane (i.e., Tom20) and endoplasmic reticulum (i.e., ERP44) proteins in the absence of Triton X-100 (Fig. 1a). Using highly purified mitochondria, we performed the nuclease protection assay with a transfected fluorescent siRNA, and found the siRNA was resistant to treatment with RNase T1 plus Micrococcal Nuclease (MNase) in the absence of Triton X-100 and became sensitive to these nucleases in the presence of the detergent (Fig. 1b). Furthermore, to visualize whether transfected siRNA distributes in mitochondria, we transfected mouse embryonic fibroblast (MEF) cells expressing TOM20-mCherry (marks the outer membrane) and su9-BFP (mitochondrial presequence of subunit 9 of the F0-ATPase which marks mitochondrial matrix) with FAM-labeled COXI siRNA. FAM signal located in su9-BFP-labeled compartment indicates the distribution of siRNA in mitochondrial matrix (Supplementary information, Fig. S1 and Video S1). These findings provide initial evidence for the ability of exogenous siRNA to localize in mitochondrial matrix.
a The purity of isolated mitochondria analyzed by western blotting for HSP60 and MRPL44 (mitochondrial matrix proteins), Tom20 (mitochondrial outer membrane protein), and ERP44 (endoplasmic reticulum protein). b The distribution of Cy3-labeled siRNA 4 h after transfection into C2C12 cells. Purified mitochondria were subjected to the nuclease protection assay with RNase T1 plus MNase. RNAs were resolved by urea or agarose gel and developed by Typhoon scanning or EtBr staining. c 32P- or Cy3-labeled RNA molecules of indicated lengths were analyzed by nuclease protection assay after incubation with purified mouse heart mitochondria. d Schematic diagram of the ClickIn assay. MitoOct carries a PPh3 group with high affinity to mitochondrial matrix and a cyclooctyne (Oct) group reactive to azide. FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; Tet, tetrazine. e Mitochondria purified from mouse heart were incubated with 32P-labeled and azide-labeled siCOXI followed by the addition of MitoOct in the presence or absence of FCCP. The upper band indicates the MitoOct-modified siRNA in mitochondrial matrix. T1, RNase T1; MN, micrococcal nuclease; MT, mitochondria.
We next used similarly purified and characterized mitochondria from mouse heart to perform an in vitro import assay with labeled RNA molecules of different lengths. After RNase treatment to remove outside RNA, we found that a significant fraction of 21- and 22-nt RNA was readily detectable within the mitochondria (Fig. 1c). In our hands, small RNAs with or without the ability to target a specific mtDNA transcript were both able to be imported in this assay (data not shown), but a slightly longer RNA of 77 nt in length was not (Fig. 1c). These data suggest that small RNA might enter mitochondria through existing pore(s) on mitochondrial membrane, but unlike the ATP-dependent mitochondrial import of tRNAs demonstrated in yeast and other lower eukaryotic organisms,22,23 small RNA import appears to take place in an ATP-independent manner (Fig. 1c). At this point, the nature of such pore or the import mechanism remains a subject of ongoing investigation (see Discussion).
Because the nuclease-resistance assay does not differentiate the localization of imported small RNA in the inter-membrane space versus the matrix of mitochondria, we next sought to determine whether small RNA is able to localize in mitochondrial matrix where siRNA is supposed to function. To this end, we employed a newly developed mitochondrial ClickIn strategy, which utilizes the mitochondria-targeted cyclooctyne (MitoOct) to conjugate any peptide or RNA that carries an azido group (Fig. 1d). As MitoOct is accumulated several hundred-fold within mitochondrial matrix driven by the mitochondrial membrane potential, it will only react with azide-labeled RNA within mitochondrial matrix, as the concentration of MitoOct elsewhere would be too low to drive this reaction.24,25 Using this strategy, we performed in vitro mitochondrial import assay with an azide-labeled siRNA and then load MitoOct onto the mitochondria. We found that ~45% of this siRNA molecules reacted with MitoOct within mitochondrial matrix (Fig. 1e). A key control of this assay is to show the dependence of MitoOct loading on the mitochondrial membrane potential, which could be blocked by the uncoupler carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP).24,25 We thus first incubated purified mitochondria with RNA, after washing the mitochondria to get rid of extra RNA, we tested the reaction of MitoOct with imported small RNA in the presence or absence of FCCP. As expected, FCCP effectively prevented this reaction (Fig. 1e). These data demonstrate the structural integrity of our purified mitochondria and the ability of small RNA to enter mitochondrial matrix.
MitoRNAi functions effectively at the RNA level
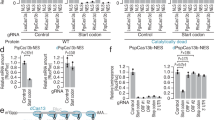
The ability of a synthetic small RNA to enter mitochondrial matrix prompted us to ask whether such small RNA could elicit RNAi within the mitochondria. We thus prepared siRNAs to target a specific mtDNA-encoded transcript (i.e., ND1). By northern blotting, we found that some siRNAs showed no effect while others were able to downregulate the ND1 transcript to a measurable degree (Supplementary information, Fig. S2a). As arbitrarily designed siRNAs seemed not to always work (which might explain early unsuccessful attempts in testing potential RNAi in mitochondria), we reasoned that the failed or inefficient cases might reflect the inaccessibility of targeted mRNA regions to those siRNAs. We thus took advantage of an atlas of Ago2-binding sites in C2C12 cells9 created earlier (by cross-linking immunoprecipitation sequencing or CLIP) to design a new set of siRNAs according to Ago2-binding peaks wherever possible (Fig. 2a; all siRNAs listed in Supplementary information, Table. S1), and observed that the new set of siRNAs each reduced the corresponding mitochondrial transcript as detected by RT-qPCR (Fig. 2b; all primers listed in Supplementary information, Table S2). We did not observe any specific sequence/motif among these effective siRNAs. To rule out the contribution of pseudogene-derived RNA to the observed effect of mitoRNAi, we generated mitochondrial DNA-deleted C2C12 cells (ρ0), then quantified mitochondrial and potential pseudogene-derived RNA. The results showed that these barely detectable transcripts were comparable to the low-level mtDNA in ρ0 cells (Fig. 2c). We further validated the siRNA effects by northern blotting in the cases of ND1 and COXI in C2C12 cells (Fig. 2d, e) and ND1 in HeLa cells (Fig. 2f). To get rid of cytosolic RNA contamination, purified mitochondria from COXII, COXIII, and ATP6 siRNA-treated C2C12 cells was analyzed by northern blotting, and these results strengthen the effect of siRNA in mitochondria (Supplementary information, Fig. S3). We further extended these findings to multiple other cell types (Supplementary information, Fig. S2b, c).
a siRNAs designed to target individual mtDNA-encoded transcripts according to previously mapped Ago2-binding peaks in C2C12 cells. b RT-qPCR analysis of all 13 mitochondrial transcripts in C2C12 cells after treatment with corresponding siRNAs. c qPCR analysis of mtDNA copy numbers in ρ0 and WT C2C12 cells (left panel); RT-qPCR analysis of transcripts in ρ0 and WT C2C12 cells (right panel). d–f Northern blot (NB) analysis of mouse ND1 and COXI mRNAs in C2C12 cells (d, e) and human ND1 mRNAs in HeLa cells (f) treated with either control or specific siRNAs. rRNAs serve as loading control and were detected by northern blotting or direct SYBRTM Gold staining in the same gel. g–j RT-qPCR analysis of COXI, II, III, and ND1 RNA levels in HEK293T cells transiently transfected (Trans.) or stably expressed (Infect.) shCOXI, shCOXII, shCOXIII, and shND1. Data represent means ± SEM based on three independent experiments. ***P < 0.005, ****P < 0.001 based on unpaired Student’s t-test.
In the cytoplasm, both transfected siRNA and plasmid-expressed shRNA can elicit RNAi. We thus tested multiple shRNAs against mtDNA-encoded ND1, COXI, COXII, and COXIII in transfected HEK293T cells and detected recordable, but somewhat variable effects (likely due to different transfection efficiencies), which became enhanced upon selection for the transfected plasmids (Fig. 2g–j). The RNAi effects observed on these individual cell lines also helped to argue against a possibility that transfection reagent might somehow stress the cells to enable transfected siRNA molecules to enter the mitochondria. Collectively, these data strongly suggest that the RNAi pathway operates in the mitochondria.
Ago2-dependent mitoRNAi
To demonstrate the targeting specificity of transfected siRNAs, we showed that siND1 targeted ND1 but not COXI, and siCOXI reduced COXI but not ND1 in two different mouse cell types (Fig. 3a). To further validate the specificity, we compared the effects of mutations (from position 9 to 13) in wild-type (WT) siCOXI and siND1 (Fig. 3b) and found that both mutant siRNAs lost the effect on their target mRNAs (Fig. 3c). This is consistent with the known base-paring requirement within the mRNA:siRNA duplex for the siRNA effect in the cytoplasm.26
a RT-qPCR analysis of ND1 or COXI in 4T1 (upper panel) and LLC (lower panel) mouse cells treated with siND1 or siCOXI. b Mutations in siCOXI and siND1 (blue) relative to WT siRNAs (red). c RT-qPCR analysis of COXI or ND1 mRNA in C2C12 cells treated with corresponding WT and mutant siRNAs. d, e WT and Ago2 KO MEFs transfected with siCOXI (d) or siND1 (e). RNAi effects were examined in Ago2 KO MEFs complemented with either a mitochondria-targeted Ago2 (su9-Ago2) or its catalytically dead D597A mutant.
The RNAi pathway is known to function via the RISC complex in the cytoplasm. In mammalian cells, Ago2, which is the only catalytically active Argonaute,8 is able to elicit the siRNA effect through its slicing activity. Using the standard curve generated with purified Ago2 protein, we quantified endogenous Ago2 protein level in purified mitochondria and the whole cell. The result indicated that about 5% of Ago2 proteins localize in mitochondria (Supplementary information, Fig. S4a). Furthermore, overexpressing mitochondria-targeted Ago2 in MEF cells enhanced the effect of mitoRNAi at RNA level, but not at protein level (Supplementary information, Fig. S4b). To determine the functional requirement of Ago2 for the newly observed RNAi activity, we tested a pair of WT and Ago2 knockout (KO) MEF cell lines. As expected, both siND1 and siCOXI downregulated their targets in WT, but not in Ago2 KO MEFs (Fig. 3d, e). The RNAi effects could be fully rescued by expressing WT Ago2, which can function in both cytoplasm and mitochondria (Supplementary information, Fig. 4c). To avoid potential compound effects due to some unknown crosstalk between the cytoplasm and the mitochondria, we tested and showed the rescue effect with a mitochondria-targeted WT Ago2 (su9-Ago2), and importantly, the slicing defective mutant su9-Ago2 (D597A) failed to rescue the RNAi activity in both siND1- and siCOXI-treated cells (Fig. 3d, e). Together, these data established Ago2 slicing activity-dependent mitoRNAi.
Impact of mitoRNAi on mtDNA-encoded proteins
We initially intended to demonstrate the RNAi effect at the protein levels, but in most cases, we saw little impact, which might have discouraged early efforts in assessing potential mitoRNAi. We reasoned that mtDNA-encoded proteins might be too long-lived, so that the RNAi effect could not be detected within the standard time window (2 to 3 days) normally used for targeting nDNA-encoded gene products. To test this possibility, we analyzed the protein levels of multiple subunits of OXPHOS complexes at different time points after blocking mitochondrial translation with chloramphenicol (INN).27 We found that representative components of OXPHOS complexes I, II, III and V all showed little change in the absence of mitochondrial translation for up to 48 h in MEF cells (Fig. 4a; all antibodies listed in Supplementary information, Table S3), and we observed similar results in C2C12 cells (Supplementary information, Fig. S5a). The only exception is complex IV in which all three mtDNA-encoded components (COXI, II, and III) and representative nDNA-encoded subunits (e.g., COX5B) were significantly reduced in 24 h after INN treatment (Fig. 4a). Corroborating these observations, the siRNAs against COXI, II, and III diminished their targets at both RNA (Supplementary information, Fig. S5b) and protein levels (Fig. 4b) in C2C12 cells. In contrast, siND1 showed little effect in reducing ND1 protein (Fig. 4c).
a Time-course analysis of representative nDNA- and mtDNA-encoded mitochondrial proteins by western blotting in MEF cells after treatment with chloramphenicol (INN). HSP60 and β-actin serve as loading controls. b Western blot analysis of COXI, COXII, or COXIII proteins after control (Ctl) or specific siRNA treatment. c Western blot analysis of ND1 protein after treatment with indicated siRNAs. d Mitochondrial translation products were pulse-labeled for 1 h in C2C12 cells treated with control or COXI siRNA for 48 h. Coomassie blue-stained gels were used to verify even loading (right panel).
We reasoned that if mitoRNAi was masked by extraordinary stability of most mtDNA-encoded proteins, compromised protein stability would enable us to detect such effects. We therefore tested knockdown of an nDNA-encoded complex I component NDUFA13, which is known to destabilize this complex,28 and indeed, we were able to record siND1 effect enhanced to a measurable degree (Fig. 4c). Furthermore, to test whether the other relatively stable complex subunit can be downregulated by siRNA, we extended the time of siRNA treatment against ND1 (CXI), CytB (CXIII), and ATP6 (CXV) from 48 to 72 h, and the results showed that these protein level decreased at the 72 h (Supplementary information, Fig. 5c). Because transcription and translation are coupled in mitochondria,29,30,31 we predicted that reduced mRNA would directly decrease the rate of nascent protein synthesis on mitochondrial ribosomes. To test this, we performed 35S-labeling experiments in the presence of Emetine to block cytoplasmic translation in C2C12 cells. As expected, COXI nascent protein synthesis was markedly reduced in response to siCOXI treatment, while translation of all other mitochondrial transcripts was unaltered (Fig. 4d). Collectively, these data provide strong evidence for active mitoRNAi, which likely escaped previous detection because of remarkable stability of most mtDNA-encoded proteins.
Using mitoRNAi to test communication among respiratory chain complexes
The establishment of active mitoRNAi afforded us a new tool to directly address some fundamental questions in mitochondrial biology, which had been mainly studied through manipulating nDNA-encoded subunits/assembly factors or by analyzing mutations in mtDNA-encoded genes identified in patients.32 To illustrate the power of this new tool, we focused on addressing potential crosstalk between different respiratory chain complexes, as various contradictory results had been reported in literature. The OXPHOS complex is composed of five complexes (I, II, III, IV, and V). Except complex II, which is only composed of nDNA-encoded subunits, all the other complexes contain both nDNA- and mtDNA-encoded subunits. Complex I, III, and IV are further assembled into various supercomplexes, and to date, the assembly of complex II has been thought to be largely independent of all other OXPHOS complexes.33,34,35 In contrast, the effect of mutations in mtDNA-encoded complex IV subunits is still under debate. Such mutations have been reported to affect complex IV only,36 to compromise both complex I and IV,37 or to impair all OXPHOS respiratory chain complexes.38,39 Because these reports are all based on analysis of patient-derived cell lines, it remains unclear whether the discrepancies result from direct impact of different mutations and/or differential adaptation of those patient-derived cells after long-term in vitro culture.
To address these possibilities, we took advantage of the newly developed siRNA tool to transiently knock down COXI, II or III in C2C12 cells, respectively, and found that they all led to compromised oxygen consumption, similar to the effect induced by downregulating nDNA-encoded COX4, as indicated by the dramatic reduction of the maximal respiratory capacity (Fig. 5a). As expected, knockdown of either COXI or COX4 reduced mitochondrial membrane potential as measured by accumulated tetramethylrhodamine methyl ester (TMRM) signal (Fig. 5b). These data corroborated the selective decrease of complex IV assembly, as revealed by blue native gel electrophoresis (BN-PAGE) (Fig. 5c) and in-gel activity assay (Fig. 5d). Using these assays, we next examined the impact of compromised complex IV on the assembly/stability of other OXPHOS complexes. We found little impact on complex I, III and V, and did not detect any effect of COX4 knockdown on these complexes either (Fig. 5e). These findings favor the possibility that the postulated influence of complex IV on the stability/assembly of complex I37 or all other respiratory chain complexes38,39 likely resulted from adaptation of patient-derived cells. Unexpectedly, we detected an increase in complex II (Fig. 5e). We speculate that compromised complex IV might reduce efficient electron transfer along the respiratory chain, thus causing a selective “damping” effect, which may account for increased complex II.
a Oxygen consumption rate (OCR) measured in C2C12 cells treated with the indicated siRNAs. COX4 serve as a positive control. Oligo Oligomycin, AA&R Antimycin A plus Rotenone. b Membrane potential measured with TMRM fluorescence in C2C12 cells treated with the indicated siRNAs. c Mitochondria were solubilized with DDM and analyzed by BN-PAGE and immunoblotting. Quantitative data were shown in the lower panel. d Mitochondria were solubilized and analyzed by BN-PAGE followed by in-gel activity assay for complex I (top) or complex IV (bottom). e BN-PAGE and immunoblotting analysis of individual mitochondrial complexes (labeled on top in each panel) with antibodies after treatment with the indicated siRNAs. The quantitative data shown in the lower panels were based on three independent biological replicates in each case.
Discussion
The reason why mitoRNAi might have escaped early detection is likely due to (i) the theoretical consideration of the bacterial origin of the mitochondria, (ii) inaccessibility of many mRNA regions to siRNAs because mitochondrial transcription and translation both take place on inner membrane, and (iii) remarkable stability of most mtDNA-encoded proteins. By experimentally addressing the latter two obstacles, we now demonstrate active RNAi in the mitochondria. Our findings are in line with our previous report of miRNA function in the mitochondria.9 Interestingly, imperfect base-pairing between a specific miRNA and its mtDNA-encoded transcript leads to enhanced translation, whereas perfect base-pairing between a specific siRNA and its target triggers mRNA degradation. While both of these reactions require Ago2, the function of siRNA, but not that of miRNA, depends on the slicing activity of Ago2.
An unsolved question is how siRNA and miRNA may translocate to the matrix of mitochondria. During this study, we have made elaborate efforts in exploring potential mechanism(s) of small RNA import into mitochondria, including targeting various molecules previously implicated in RNA import into mitochondria, such as PNPT1,40 GRSF1,41 and the TOM complex.42 We also tested blockage of mitochondrial outer membrane permeability (MOMP) and the permeability transition pore (MPTP).43,44 So far, we have not yet obtained any convincing data. Of note, a recent report suggests that mitochondrial RNA export plays a role in inducing innate immunity in mammalian cells,45 but the mechanism also remains elusive. Therefore, future studies are clearly required to elucidate the mechanism of RNA import to or export from mitochondria. We suggest that active mitoRNAi may serve as a functional readout for future investigation of this fundamental problem.
Despite unclear mechanism underlying the entrance of small RNA to mitochondria, the established activities of both siRNA and miRNA within the mitochondria would provide very useful tools to directly explore the function of mtDNA-encoded genes, which has been solely relying on patient-derived cells. It is interesting to note that miRNA-enhanced mitochondrial translation has been linked to a role of miR-21 in lowing blood pressure in a rat spontaneous hypertension model.46 The newly demonstrated mitoRNAi provides a potential therapeutic intervention strategy against human diseases caused by mutations in the mitochondrial genome. For example, this precise RNA cleavage tool may be used to selectively remove mutated mitochondrial transcripts, which often co-exist with WT counterparts in patients with heteroplasmic mtDNA mutations. Thus, the newly established mitoRNAi is expected to drive both basic and clinical research on mitochondrial biology.
Materials and methods
Cell culture and transfection
C2C12, LLC, MEF, 4T1, HeLa, and 293 T cells were all cultured in Dulbecco’s Minimal Essential Medium (DMEM, Life Technology) supplemented with 10% fetal bovine serum (FBS, Life Technology). Cells were maintained in 5% CO2 at 37 °C. For transfection with siRNA, Lipofectamine RNAiMAX (Invitrogen) was used according to the manufacturer’s instruction. After siRNA treatment for 48 h, cells were collected with Trizol (Invitrogen) for RNA analysis by RT-qPCR and northern blotting or with SDS loading buffer for protein analysis by western blotting.
Generation of shRNA-expressing stable cell lines
Synthetic shRNA oligonucleotides were inserted into the pLKO.1 plasmid between AgeI and EcoRI sites. The plasmid was co-transfected with psPAX2 and pMD2G at a 5:3.75:1.25 ratio into HEK293T cells to produce lentivirus in 6 cm dish. Collected lentivirus was used to infect HEK293T for 24 h followed by selection using puromycin (Sigma) at the concentration of 2 μg/mL for 6 days.
Mitochondria isolation
Mouse heart or cells were homogenized with isolation buffer (250 mM Sucrose, 10 mM Tris-HCl, pH 7.4, 1 mM EDTA) by Dounce (Sigma), as described earlier.47 The homogenate was next separated with the following steps: first, clear cell debris at 800× g twice and then at 1200× g twice in 4 °C for 5 min; second, transfer the supernatant to a new tube and pellet crude mitochondria at 15,000× g for 10 min; third, resuspend the crude mitochondria pellet with isolation buffer and lay it on a sucrose gradient (17%, 31%, 42%, 50%) prepared with T10E20 buffer (10 mM Tris-HCl, pH 7.4 and 20 mM EDTA-Tris, pH 7.4), as previously described.9 The sucrose gradient with crude mitochondria was centrifuged in SW41 rotor (Beckman) at 18,000 rpm for 45 min. Mitochondria were collected at the layer between 42% and 50% sucrose.
Small RNA importing assay
To assay for small RNA import in vitro, mitochondria isolated from mouse heart were divided into five aliquots. Individual aliquots were incubated with 77-nt 32P-labeled RNA or 22-nt Cy3-labeled RNA at 37 °C for 30 min. Two of these aliquots were also added with ATP (2 mM). After incubation, the rest of aliquots except the input aliquot were subjected to RNase T1 (3000 U/mL, Thermo Scientific) digestion on ice for 30 min. Mitochondria were pelleted by centrifugation and RNA was extracted with Trizol. RNA in aliquots incubated with 77-nt 32P-labeled RNA were resolved in a 12% urea-page gel whereas RNA in aliquots incubated with 22-nt Cy3-labeled RNA were resolved in a 15% urea-page gel. The gels were scanned on a typhoon instrument (GE) for quantification.
To assay for small RNA import in vivo, mitochondria isolated from C2C12 cells transfected with 22-nt Cy3-labeled siRNA were divided into three aliquots with one aliquot standing on ice as control, and the other two subjected to digestion with RNase T1 (3000 U/mL) plus Micrococcal nuclease (1000 U/mL) on ice for 30 min in the presence or absence of Triton X-100, as described earlier.9 Total lysate served as control. Mitochondria were pelleted by centrifugation at 20,000× g and RNA was extracted with Trizol for analysis by RT-qPCR. 7SK, 16S rRNA, and GAPDH RNAs served as internal controls.
ClickIn chemistry assay
The ClickIn chemistry assay was performed as reported.24 Briefly, purified mitochondria from mouse heart were divided into 3 aliquots and all incubated with 32P-labeled and azido-tagged siRNA in KCl buffer (120 mM KCl, 10 mM HEPES-KOH, 1 mM EGTA, 1 mM ADP, 1 mM MgCl2, 1 mM KPi, 0.05% BSA, pH 7.4 supplemented with 10 mM potassium succinate) for 1 h. Free RNA was removed by washing with STE buffer (250 mM sucrose, 5 mM Tris-HCl, 1 mM EGTA, pH 7.4) for 3 times. MitoOct was added in the presence or absence of FCCP and incubated at 37 °C for 1 h. After incubation, mitochondria were pelleted at 12,000× g for 2 min, and after washing with STE buffer, the pellet was resuspended in 50 μL STE buffer. RNase T1 and Tetrazine were added at the final concentrations of 3000 U/mL and 50 μM, respectively, and the reactions were incubated on ice for 20 min. Mitochondria were pelleted by centrifugation at 20,000× g, and RNA was extracted with Trizol for downstream analysis. RNA-azide-Oct could be readily separated from unreacted RNA-azide in 20% urea denaturing gel. The gel was exposed to a phosphor imager overnight and scanned in a Typhoon Scanner (GE).
Northern blotting and RT-qPCR
For northern blotting, C2C12 or HeLa cells were first transfected with siRNA for 48 h, and RNA was extracted with Trizol. 10 μg RNA was separated on 1.5% formaldehyde agarose gel and blotted with probes (listed in Supplementary information, Table S2). For northern blot analysis of COXI RNA after siRNA treatment, mitochondria were first purified before RNA extraction. For RT-qPCR, RNA was extracted with Trizol 48 h after siRNA transfection. cDNA was synthesized by reverse-transcription (M-MLV, Promega) with random primers (N9). cDNA was quantified with Master SYBR Green Mix (Roche) on Bio-Rad Touch CFX96.
Analysis of mitochondrial protein stability
MEFs or C2C12 cells were first plated on 6-well plate with 10% FBS DMEM, and after reaching 70%–80% confluency, the culture medium was replaced with 10% FBS DMEM containing 250 μg/mL (for MEFs) or 150 μg/mL (for C2C12 cells) chloramphenicol. Samples were collected from 0 to 48 h at 12-h intervals. All samples were lysed with SDS loading buffer and analyzed by western blotting with different antibodies (listed in Supplementary information, Table S3).
Mitochondrial translation assay
35S-Cys/Met was used to label nascent peptides according to the procedure.48 C2C12 cells were first transfected with siRNA on 6-well plate and culture medium was replaced with labeling medium (cysteine and methionine-free DMEM supplemented with 10% dialyzed FBS) for 30 min at 37 °C. Emetine (100 μg/mL) was added to the labeling medium for 5 min. EasyTag 35S-labeling mixture (PerkinElmer) was added to the labeling medium to final 200 μCi/mL and the reaction was incubated for 1 h. Labeled proteins were collected with SDS loading buffer and resolved in 17.5% SDS-PAGE. After electrophoresis, the gel was dried and exposed to phosphor screen for 2 days and analyzed on a Typhoon scanner (GE).
BN-PAGE and in-gel activity
Mitochondrial respiratory complex abundance and activity were analyzed with BN-PAGE and in gel activity assay, respectively. The procedure of BN-PAGE was as previously described.49 Briefly, C2C12 cells were first transfected with siRNA and plated on 60 mm dish. After cell confluency reaching 90% in ~48 h, cells were transferred to a 1.5 mL tube and washed once with ice-cold PBS. Cells were pelleted and protein concentration was analyzed with BCA (Pierce). The cell pellet was resuspended with a volume (X) of ice-cold PBS to reach a final concentration of 5 mg/mL. An equal volume of digitonin (4 mg/mL) was added and the reaction was incubated on ice for 10 min. The pellet was washed three times with pre-cold PBS to remove residual digitonin and then resuspended with BN sample buffer (half volume of X) and 10% DDM (one-tenth volume of X) and then incubated on ice for 20 min. Sample was cleared by centrifugation at 20,000× g for 20 min, and the supernatant was transferred to a fresh tube containing 5% SBG (half volume of 10% DDM used). BN samples were separated on 3%–15% blue native gel. Antibodies were used to probe the complex abundance (listed in Supplementary information, Table S3). Reagents used to detect complex I and complex IV activities were as described.50
Measurement of mitochondrial oxygen consumption rate and membrane potential
Mitochondrial oxygen consumption rate was measured on Seahorse as described.51 C2C12 cells were first transfected with siRNA in 12-well plate. After cell confluency reaching 90% in 48 h, about 30,000 cells were replated on a 24-well Seahorse plate. Cells were refreshed with detection solution gradually to the final volume of 525 μL and incubated at 37 °C for 1 h. Cells were configured with a mito stress kit (Agilent Technologies) and probe plate pretreated with XF Celebrant pH 7.4 at 37 °C according to the manufacturer’s instruction. The samples were analyzed with Seahorse XF24 software on Seahorse XF24 Extracellular Flux Analyzer (Agilent Technologies).
Mitochondrial membrane potential was measured by FACS (fluorescence-activated cell sorting), as described.52 C2C12 cells were first transfected with siRNA and seeded on 6-well plate. Trypsinized cells were resuspended in DMEM and washed twice with pre-cold PBS. Cells were stained with 100 μL 100 nM TMRM (Invitrogen) in DMEM without phenol red and incubated at 37 °C for 30 min. Stained cells were washed twice with pre-cold PBS and detected by FACS on the exciting wavelength 488 nm and emission wavelength 570 nm. Data were analyzed with FlowJo.
Immunocytochemistry
FAM-conjugated COXI siRNAs were transfected into MEF cells expressing TOM20-mCherry (outer membrane) and subunit 9 of ATP synthase (su9)-BFP (matrix). Five hours after transfection, cells were washed with PBS 5–10 times and analyzed with Structured Illumination Microscopy (SIM) (Delta Vision OMX). The signals captured by SIM were reconfigured with the Softworx 6.0 Beta 19. Then the 3D image was processed with Imaris. The FAM-siCOXI signal in mitochondria was extracted and the signal outside of mitochondria was discarded. The picture and movie were all produced by Imaris.
References
Drinnenberg I. A. et al. RNAi in budding yeast. Science 326, 544–550 (2009).
Fire A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Hamilton, A. J. & Baulcombe, D. C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952 (1999).
Imig J. et al. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction. Nat. Chem. Biol. 11, 107–114 (2015).
Elbashir S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Shi Y. Mammalian RNAi for the masses. Trends Genet. 19, 9–12 (2003).
Roger, A. J., Munoz-Gomez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Liu J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004).
Zhang X. R. et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158, 607–619 (2014).
Kren B. T. et al. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 6, 65–72 (2009).
Koopman, W. J. H., Distelmaier, F., Smeitink, J. A. M. & Willems, P. H. G. M. OXPHOS mutations and neurodegeneration. EMBO J. 32, 9–29 (2013).
Ojala, D., Montoya, J. & Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474 (1981).
Moulin, C., Caumont-Sarcos, A. & Ieva, R. Mitochondrial presequence import: Multiple regulatory knobs fine-tune mitochondrial biogenesis and homeostasis. Biochim. Biophys. Acta Mol. Cell Res. 1866, 930–944 (2019).
Jeandard D. et al. Import of non-coding RNAs into human mitochondria: a critical review and emerging approaches. Cells 8, 286 (2019).
Isaac, R. S., McShane, E. & Churchman, L. S. The multiple levels of mitonuclear coregulation. Annu. Rev. Genet. 52, 511–533 (2018).
Schneider A. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int. J. Parasitol. 31, 1403–1415 (2001).
Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 80, 1033–1053 (2011).
Holzmann J. et al. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135, 462–474 (2008).
Brown A. et al. Structure of the large ribosomal subunit from human mitochondria. Science 346, 718–722 (2014).
Kiss, T. & Filipowicz, W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell 70, 11–16 (1992).
Das S. et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 110, 1596–1603 (2012).
Mahapatra, S., Ghosh, T. & Adhya, S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 22, 3381–3386 (1994).
Rubio M. A. et al. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Natl. Acad. Sci. USA. 105, 9186–9191 (2008).
Hoogewijs K. et al. ClickIn: a flexible protocol for quantifying mitochondrial uptake of nucleobase derivatives. Interface Focus 7, 20160117 (2017).
Logan A. et al. Assessing the mitochondrial membrane potential in cells and in vivo using targeted click chemistry and mass spectrometry. Cell Metab. 23, 379–385 (2016).
Wang Y. et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926 (2008).
McKee, E. E., Ferguson, M., Bentley, A. T. & Marks, T. A. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrobial Agents Chemother. 50, 2042–2049 (2006).
Stroud D. A. et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538, 123–126 (2016).
Kehrein K. et al. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 10, 843–853 (2015).
Bogenhagen, D. F., Martin, D. W. & Koller, A. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 19, 618–629 (2014).
Rackham O. et al. Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep. 16, 1874–1890 (2016).
Taylor, R. W. & Turnbull, D. M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6, 389–402 (2005).
Gu J. et al. The architecture of the mammalian respirasome. Nature 537, 639–643 (2016).
Wu, M., Gu, J., Guo, R., Huang, Y. & Yang, M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167, 1598–1609 (2016).
Guo, R., Zong, S., Wu, M., Gu, J. & Yang, M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170, 1247–1257 (2017).
Tiranti V. et al. A novel frameshift mutation of the mtDNA COIII gene leads to impaired assembly of cytochrome c oxidase in a patient affected by Leigh-like syndrome. Hum. Mol. Genet. 9, 2733–2742 (2000).
Li Y. et al. An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem. 282, 17557–17562 (2007).
Rahman S. et al. A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am. J. Hum. Genet. 65, 1030–1039 (1999).
Hornig-Do H. T. et al. Nonsense mutations in the COX1 subunit impair the stability of respiratory chain complexes rather than their assembly. EMBO J. 31, 1293–1307 (2012).
Wang G. et al. PNPASE regulates RNA import into mitochondria. Cell 142, 456–467 (2010).
Noh J. H. et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 30, 1224–1239 (2016).
Wiedemann, N. & Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 86, 685–714 (2017).
Schinzel A. C. et al. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc. Natl. Acad. Sci. USA. 102, 12005–12010 (2005).
Halestrap A. P. What is the mitochondrial permeability transition pore? J. Mol. Cell Cardiol. 46, 821–831 (2009).
Dhir A. et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018).
Li H. et al. MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation. Circulation 134, 734–751 (2016).
Frezza, C., Cipolat, S. & Scorrano, L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2, 287–295 (2007).
Sasarman, F. & Shoubridge, E. A. Radioactive labeling of mitochondrial translation products in cultured cells. Methods Mol. Biol. 837, 207–217 (2012).
Leary S. C. Blue native polyacrylamide gel electrophoresis: a powerful diagnostic tool for the detection of assembly defects in the enzyme complexes of oxidative phosphorylation. Methods Mol. Biol. 837, 195–206 (2012).
Khvorostov, I., Zhang, J. & Teitell, M. Probing for mitochondrial complex activity in human embryonic stem cells. J. Vis. Exp. 17, 724 (2008).
Gao Y. et al. Mammalian elongation factor 4 regulates mitochondrial translation essential for spermatogenesis. Nat. Struct. Mol. Biol. 23, 441–449 (2016).
Khacho M. et al. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell 19, 232–247 (2016).
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology of China (2017YFA0504600, 2019YFA0508700, and 2017YFA0504400), the National Natural Science Foundation of China (31670825 and 91440102), and Chinese Academy of Science foundation grant (22KJZD-EW-L12) to X. Zhang. Work in the Murphy Lab was supported by the Medical Research Council UK (MC_U105663142) and by a Wellcome Trust Investigator award (110159/Z/15/Z). Work of K.H. is funded by the FWO (2018, 1.5.193.18N) and EU Horizon 2020 research and innovation program (665501). We also acknowledge the contribution of Drs. Yi Zhang and Yuanchao Xue in the early phase of this project, Dr. Heping Cheng for providing the Seahorse apparatus, and Hongjie Zhang for isotope experiment assistance. We are grateful to Drs. Immo Scheffler, Jing Hu, and Peter Rehling for critical comments on the manuscript.
Author information
Authors and Affiliations
Contributions
X. Zhang and X.-D.F. designed the experiments; K.G., M.C., and X. Zhang performed most experiments; X. Zuo helped with data analysis; J.L. contributed to experimental design and data interpretation; M.P.M. and K.H. designed the ClickIn experiment and provided the MitoOct reagents; X. Zhang, K.G., and X.-D.F. wrote the manuscript; and M.P.M. and K.H. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Gao, K., Cheng, M., Zuo, X. et al. Active RNA interference in mitochondria. Cell Res 31, 219–228 (2021). https://doi.org/10.1038/s41422-020-00394-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-020-00394-5
This article is cited by
-
mcPGK1-dependent mitochondrial import of PGK1 promotes metabolic reprogramming and self-renewal of liver TICs
Nature Communications (2023)
-
Photons guided by axons may enable backpropagation-based learning in the brain
Scientific Reports (2022)
-
Neddylation regulation of mitochondrial structure and functions
Cell & Bioscience (2021)