Abstract
Type I interferon (IFN-I) production is efficiently induced to ensure a potent innate immune response to viral infection. How this response can be enhanced, however, remains to be explored. Here, we identify a new cytoplasmic long non-coding RNA (lncRNA), lncLrrc55-AS, that drives a positive feedback loop to promote interferon regulatory factor 3 (IRF3) signaling and IFN-I production. We show that lncLrrc55-AS is virus-induced in multiple cell types via the IFN-JAK-STAT pathway. LncLrrc55-AS-deficient mice display a weakened antiviral immune response and are more susceptible to viral challenge. Mechanistically, lncLrrc55-AS binds phosphatase methylesterase 1 (PME-1), and promotes the interaction between PME-1 and the phosphatase PP2A, an inhibitor of IRF3 signaling. LncLrrc55-AS supports PME-1-mediated demethylation and inactivation of PP2A, thereby enhancing IRF3 phosphorylation and signaling. Loss of PME-1 phenocopies lncLrrc55-AS deficiency, leading to diminished IRF3 phosphorylation and IFN-I production. We have identified an IFN-induced lncRNA as a positive regulator of IFN-I production, adding mechanistic insight into lncRNA-mediated regulation of signaling in innate immunity and inflammation.
Similar content being viewed by others
Introduction
The innate immune response is a host’s first line of defense against invading viruses.1 Innate receptors recognize viral components and activate downstream signaling, including the interferon regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB) pathways.2,3 Activation of the transcription factor IRF3 induces production of type I interferon (IFN-I),4 which plays a critical role in host defense against viral invasion.5 Upstream signaling molecules (e.g., TBK1, IKKε) phosphorylate IRF3, triggering its dimerization and translocation to the nucleus, where it stimulates IFN-I expression.6 Protein Phosphatase 2A (PP2A) dephosphorylates IRF3 and thus negatively regulates IFN-I production.7 How phosphorylation and dephosphorylation of IRF3 are precisely balanced to ensure appropriate IFN-I production while avoiding tissue damage during the innate immune response is not fully understood.
Epigenetic regulators are known to play important roles in the regulation of immune cell functions and also in the pathogenesis of immune disorders.8,9,10,11 Long non-coding RNAs (lncRNAs) are emerging as critical regulators of both innate and adaptive immunity. Some lncRNAs directly interact with chromatin-modifying factors, heterogeneous nuclear ribonucleoproteins (hnRNPs), or transcription factors to regulate the transcription of immune-related genes, whereas others form multi-subunit complexes to regulate innate immune response pathways.12,13,14,15,16 Cytoplasmic lncRNAs have been shown to control the activity of signaling components by modulating post-translational modifications (PTMs) or cellular metabolism.17,18 In addition, some differentially expressed lncRNAs regulate inflammatory innate responses and pathogen evasion or survival during host-pathogen interactions.17,19,20,21 LncRNAs can also interact with signaling molecules to control diverse biological processes.
We previously reported that viral infection induces expression of the IFN-I-dependent lncRNA lnc-lsm-3b and the IFN-I-independent lncRNA lncRNA-ACOD1, which regulate the innate response and viral replication via different mechanisms.17,20 To identify lncRNAs that can selectively regulate antiviral IFN-I induction, we examined the function of lncRNAs that are upregulated in virus-infected macrophages. We discovered a new IFN-I-inducible lncRNA, lncLrrc55-AS, that selectively promotes antiviral IFN-I production by strengthening IRF3 phosphorylation.
Results
Identification of lncLrrc55-AS as an IFN-I-inducible lncRNA
We previously identified several IFN-independent lncRNAs that were upregulated upon vesicular stomatitis virus (VSV) infection in wild-type (WT) macrophages and IFNα/β receptor knockout macrophages.17 We also identified several lncRNAs that are IFNα/β receptor dependent.17 RNA interference (RNAi)-mediated silencing of several of these lncRNAs led to higher VSV titers in the supernatant of infected macrophages (Supplementary information, Fig. S1a). Of these lncRNAs, the transcript annotated as ENSMUST00000155472.7, which maps to chr2qD (chr2:85,160,778–85,173,936, mm10), caught our attention. We named this transcript lncLrrc55-AS because it is derived from the antisense transcripts of the Lrrc55 (leucine rich repeat containing 55) gene (Supplementary information, Fig. S1b). LncLrrc55-AS is a spliced transcript of 286 nucleotides (nt) with a 3′ polyadenylated (poly A) tail, as indicated by RT-PCR and RACE (rapid amplification of cloned cDNA ends) (Fig. 1a; Supplementary information, Fig. S1c, Table S2). Infection of mouse peritoneal macrophages with another RNA virus, Sendai virus (SeV), also triggered upregulation of lncLrrc55-AS (Fig. 1b).
Identification of virus-induced IFN-I-dependent lncLrrc55-AS in macrophages. a cDNA of lncLrrc55-AS in VSV-infected mouse peritoneal macrophages. b RT-qPCR analysis of lncLrrc55-AS in macrophages infected with SeV for 12 h. c RT-qPCR analysis (left) of the distribution of different RNAs following nucleus/cytoplasm fractionation of macrophages infected with SeV for 12 h. Western blot analysis (right) of nucleus/cytoplasm fractionation. d FISH analysis of lncLrrc55-AS in macrophages infected with SeV for 12 h. Scale bar, 20 µm. e, f RT-qPCR analysis of lncLrrc55-AS expression in macrophages infected with HSV for 12 h, or stimulated with poly(I:C) for 12 h, LPS for 3 h (e), or IFNβ with the indicated dose for 8 h (f). g, h RT-qPCR analysis of lncLrrc55-AS expression in WT and Irf3−/− macrophages infected with SeV or VSV for 12 h (g), in WT and Ifnar1−/− macrophages treated with IFNα4, IFNβ (500 U/mL) for 8 h or infected with SeV for 12 h (h). i RT-qPCR analysis of lncLrrc55-AS expression in macrophages pretreated for 1 h with the JAK inhibitor Baricitinib (2 μM) or the STAT1 inhibitor Fludarabine (1 μM), then infected with SeV or VSV for 12 h or treated with IFNα4 for 8 h. Data are from three independent experiments (b, c left, e–i, means ± SEM) or are representative of three independent experiments with similar results (a, c right, d). ***P < 0.005 (Student’s t-test or ANOVA)
Bioinformatic analysis showed that lncLrrc55-AS lacks coding potential, with a PhyloCSF score < 0 according to NCBI ORF finder (Supplementary information, Fig. S1d, e). Public ribosome profiling data revealed that lncLrrc55-AS has very low ribosome occupancy (Supplementary information, Fig. S1f). Moreover, lncLrrc55-AS cloned into any frame of the pCDNA3.1 (-B) Flag expression vector did not generate Flag-tagged peptides (Supplementary information, Fig. S1g). We found that lncLrrc55-AS primarily localized to the cytoplasm of infected macrophages, as determined by RT-qPCR analysis of subcellular fractions (Fig. 1c) and fluorescent in situ hybridization (FISH) (Fig. 1d). The human lncLrrc55-AS ortholog (which has yet to be functionally characterized) was predicted on chromosome 11, and shares 84% homology with mouse lncLrrc55-AS (Supplementary information, Fig. S1h). Together, these data suggest that lncLrrc55-AS is a non-coding RNA that localizes to the cytoplasm.
LncLrrc55-AS upregulation was dependent on virus dose (Supplementary information, Fig. S2a), and was triggered by various innate stimuli, including infection with a DNA virus (HSV), as well as exposure to pattern recognition receptor ligands (lipopolysaccharides (LPS), poly (I: C)) (Fig. 1e), and IFN-I (IFNα or IFNβ) (Fig. 1f; Supplementary information, Fig. S2b). Absolute copy number analysis revealed that lncLrrc55-AS was present at a low level in uninfected cells, and increased to ~50 copies per cell in both peritoneal macrophage infected with SeV or in NIH/3T3 cell infected with VSV for 12 h (Supplementary information, Fig. S2c), which is similar in abundance to other lncRNAs.13,17,20 In addition, VSV infection induced lncLrrc55-AS expression in multiple murine cell lines in vitro (Supplementary information, Fig. S2d) and multiple tissues in vivo (Supplementary information, Fig. S2e). Thus, lncLrrc55-AS is upregulated in a variety of cell types in response to innate stimuli.
To uncover the signaling pathway that induces lncLrrc55-AS expression, we performed genetic and pharmacological analyses. We found that lncLrrc55-AS was not induced in macrophages with defective IFN signaling, including IRF3-knockout (Irf3−/−) cells infected with SeV or VSV (Fig. 1g), or in IFNα/β receptor-knockout (Ifnar1−/−) cells infected with SeV or stimulated with IFN-I (Fig. 1h). The interferon-stimulated gene Isg15 was not upregulated in Ifnar1−/− cells, verifying their IFN signaling deficiency (Supplementary information, Fig. S3a). Inhibition of JAK1/JAK2 and STAT1 signaling by treating macrophages with Baricitinib or Fludarabine, respectively, abrogated the innate stimuli-induced upregulation of lncLrrc55-AS (Fig. 1i; Supplementary information, Fig. S3a). Consistent with these results, bioinformatic analysis with LASAGNA-Search software indicated the presence of STAT1 binding sites in the promoter region of lncLrrc55-AS (Supplementary information, Fig. S3b). We also found that the accumulation of IFN-I occurred earlier than the induction of lncLrrc55-AS in SeV-infected macrophages (Supplementary information, Fig. S3c). Taken together, these data confirm that lncLrrc55-AS is IFN-I inducible.
LncLrrc55-AS enhances IFN-I production both in vitro and in vivo
To investigate a potential role for lncLrrc55-AS in antiviral immunity, we examined the effects of lncLrrc55-AS depletion on infection. We found that the siRNAs did not affect Lrrc55 mRNA expression (Fig. 2a; Supplementary information, Fig. S4a). RNAi-mediated silencing of lncLrrc55-AS in macrophages enhanced VSV and HSV replication (Fig. 2b, c; Supplementary information, Fig. S4b), and also led to increased levels of influenza virus A (PR8 strain) RNA in infected macrophages (Supplementary information, Fig. S4c). These data suggested that the IFN-I-inducible lncLrrc55-AS may act as a positive regulator to promote the antiviral innate response.
LncLrrc55-AS enhances IFN-I production in response to viral infection. a RT-qPCR analysis of lncLrrc55-AS silencing efficiency by two siRNAs (#1, #2). NC, a non-targeting control siRNA. b Fluorescence analysis of VSV replication in NC- or lncLrrc55-AS-silenced peritoneal macrophages infected with VSV-GFP for the indicated hours. c Determination of virus loads by TCID50 assay of the supernatant from macrophages infected with VSV for the indicated hours. d ELISA of IFNα and IFNβ in supernatant of control or lncLrrc55-AS-silenced macrophages infected with VSV for 12 h. e RT-qPCR analysis of lncLrrc55-AS or Ifnb mRNA expression in the selected WT or lncLrrc55-AS KO NIH/3T3 cell clones infected with VSV for 12 h. f RT-qPCR analysis of Ifna4 and Ifnb mRNA expression in empty vector control (EV Ctrl) or lncLrrc55-AS transfected lncLrrc55-AS KO NIH/3T3 cells infected with VSV for 12 h. Data are from three independent experiments (a, b right, c–f, means ± SEM) or are representative of three independent experiments with similar results (b left). *P < 0.05, **P < 0.01 and ***P < 0.005; ns, no significance (Student’s t-test or ANOVA)
We subsequently found that silencing of lncLrrc55-AS significantly impaired IFNα and IFNβ production in RNA virus-infected macrophages (Fig. 2d; Supplementary information, Fig. S4d). To further validate these findings, we used CRISPR-Cas9 technology to generate a lncLrrc55-AS knockout (KO) NIH/3T3 cell line in which the first exon of the endogenous lncLrrc55-AS gene was replaced with a GFP-poly A signal cassette (Supplementary information, Fig. S4e, f). The lncLrrc55-AS KO NIH/3T3 cells exhibited a similar proliferation capacity to WT cells (Supplementary information, Fig. S4g). The depletion of lncLrrc55-AS did not affect Lrrc55 mRNA expression (Supplementary information, Fig. S4h). LncLrrc55-AS KO NIH/3T3 cells showed diminished induction of Ifnb mRNA upon VSV infection (Fig. 2e). Importantly, ectopic expression of lncLrrc55-AS restored VSV-induced Ifna4 and Ifnb mRNA expression in lncLrrc55-AS KO NIH/3T3 cells (Supplementary information, Fig. S4i; Fig. 2f). Thus, lncLrrc55-AS can enhance IFN-I production in antiviral innate immunity.
To investigate the biological function of lncLrrc55-AS in vivo, we generated lncLrrc55-AS-deficient (lncLrrc55-AS−/−) mice, employing the same strategy that we used to generate NIH/3T3 KO cells (Supplementary information, Fig. 4e, f). These lncLrrc55-AS−/− mice did not manifest any gross developmental defects or abnormal behaviors (data not shown), nor did lncLrrc55-AS deficiency affect CD4+/CD8+ T cell, B cell, or neutrophil population distribution in the spleen (Supplementary information, Fig. S5a). When challenged with VSV, lncLrrc55-AS−/− mice displayed no lncLrrc55-AS expression, and had a significantly increased VSV titer, enhanced viral replication in various organs and elevated inflammatory cell infiltration compared to WT mice (Fig. 3a–c; Supplementary information, Fig. S5b). lncLrrc55-AS−/− mice also had lower levels of serum IFNβ after 12 h of VSV infection compared to WT controls (Fig. 3d), consistent with decreased antiviral resistance. Additionally, the induction of Ifna4 and Ifnb mRNA expression by the infection of VSV or SeV was reduced in lncLrrc55-AS−/− peritoneal macrophages compared to WT controls (Fig. 3e; Supplementary information, Fig. 6a). Finally, lncLrrc55-AS−/− mice exhibited decreased survival upon VSV infection compared to WT littermates (Fig. 3f).
LncLrrc55-AS protects mice against viral infection. a Determination of VSV loads by TCID50 in organs of WT or lncLrrc55-AS−/− mice (n = 6 per group, 7 weeks old) 12 or 18 h after intraperitoneal injection of VSV (5 × 107 pfu/g). b Determination of VSV replication by RT-qPCR in organs of WT or lncLrrc55-AS−/− mice 18 h after infection with VSV (as in a). c Hematoxylin and eosin staining of lung sections from WT or lncLrrc55-AS−/− mice 18 h after infection with VSV (as in a). Scale bar, 50 µm. d ELISA of cytokines in serum from WT or lncLrrc55-AS−/− mice 12 h after infection with VSV (as in a, n = 4). e RT-qPCR analysis of lncLrrc55-AS, Ifna4 and Ifnb mRNA expression in peritoneal macrophages from WT or lncLrrc55-AS−/− mice 12 h after infection with VSV (as in a, n = 3). f Survival of 7-week-old WT or lncLrrc55-AS−/− mice (n = 10) after intraperitoneal injection of VSV (1 × 108 pfu/g). Kaplan–Meier curve was used to evaluate survival rate. Data are shown as means ± SEM (a, b, d, e), or are representative of three independent experiments with similar results (c). *P < 0.05, **P < 0.01 and ***P < 0.005 (Student’s t-test)
Taken together, these data suggest that lncLrrc55-AS promotes IFN-I production both in vitro and in vivo, leading to efficient induction of the innate response against viral infection.
LncLrrc55-AS enhances IFN-I production via IRF3
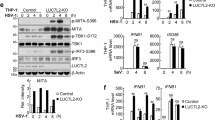
To gain insight into how lncLrrc55-AS promotes IFN-I production, we performed reporter assays in 293T cells. Briefly, we co-transfected a lncLrrc55-AS expression plasmid and an IFNβ luciferase reporter together with plasmids expressing the innate signaling components RIG-I, MAVS, TBK1 or IRF3. Ectopic expression of lncLrrc55-AS alone did not activate the IFNβ luciferase reporter, but it enhanced the activation mediated by RIG-I, MAVS, TBK1 and IRF3. In contrast, lncLrrc55-AS co-expressed with a constitutively inactive form of IRF3, IRF3-5A, did not activate the reporter (Fig. 4a; Supplementary information, Fig. S6b). We examined downstream signaling pathways and found that IRF3 phosphorylation was markedly reduced in lncLrrc55-AS-silenced macrophages after SeV infection (Fig. 4b; Supplementary information, Fig. S6c) and in lncLrrc55-AS KO NIH/3T3 cells after VSV infection (Fig. 4c). Ectopic expression of lncLrrc55-AS restored IRF3 phosphorylation in lncLrrc55-AS KO NIH/3T3 cells infected with VSV (Fig. 4d). We found that the IFN-I expression pattern and IRF3 phosphorylation levels over 24 h of viral infection, specifically, a rapid increase followed by a sharp decrease, did not correlate with lncLrrc55-AS expression, which exhibited a rapid increase followed by a plateau (Supplementary information, Fig. S3c). These results indicate that endogenous lncLrrc55-AS may affect IRF3-mediated IFN-I production with a ceiling, which aligns with previous studies indicating that IFN-I production can be mediated via several mechanisms that allow cellular homeostasis.19 To explore this possibility, macrophages ectopically expressing lncLrrc55-AS were infected with SeV for 8 h. As expected, ectopically expressed lncLrrc55-AS in WT macrophages could induce a higher level of IRF3 phosphorylation, and Ifna4 and Ifnb mRNA expression (Fig. 4e, f). However, normal expression of Ifna4 or Ifnb mRNA was not induced in Irf3−/− macrophages infected with SeV for 8 h, and this defect was not rescued by ectopic expression of lncLrrc55-AS (Fig. 4f). These results suggest that lncLrrc55-AS enhances antiviral Ifna4 and Ifnb mRNA expression by promoting IRF3 phosphorylation.
LncLrrc55-AS enhances IRF3 signaling in antiviral innate responses. a Dual luciferase analysis of the activity of the IFNβ promoter after co-transfecting plasmids expressing lncLrrc55-AS (or empty vector control, EV Ctrl), an IFNβ reporter and TK-renilla with RIG-I, MAVS, TBK1, IRF3 or IRF3-5A in 293T cells. b, c Western blot analysis of phosphorylated (p-) or total protein in lysates of NC- and lncLrrc55-AS-silenced peritoneal macrophages infected with SeV (b) or WT control and lncLrrc55-AS KO NIH/3T3 cells infected with VSV (c) for the indicated hours. d Western blot analysis of phosphorylated (p-) or total IRF3 in lysates of lncLrrc55-AS KO NIH/3T3 cells ectopically expressing EV Ctrl or lncLrrc55-AS after VSV infection for the indicated hours. e Western blot analysis of phosphorylated (p-) or total IRF3 in lysates of WT and Irf3−/− peritoneal macrophages ectopically expressing lncLrrc55-AS or EV Ctrl after SeV infection for 8 h. f RT-qPCR analysis of lncLrrc55-AS, Ifna4 and Ifnb mRNA expression of WT and Irf3−/− peritoneal macrophages ectopically expressing lncLrrc55-AS or EV Ctrl after SeV infection for 8 h. Data are from three independent experiments (a, f, means ± SEM) or are representative of three independent experiments with similar results (b–e). *P < 0.05, **P < 0.01; ns, no significance (Student’s t-test or ANOVA)
Phosphatase methylesterase 1 (PME-1) is required for lncLrrc55-AS promotion of IRF3 signaling and IFN-I production
To elucidate how lncLrrc55-AS promotes IRF3-mediated IFN-I production, we sought to identify protein partners of lncLrrc55-AS. Specifically, we used an RNA antisense purification strategy to purify endogenous lncLrrc55-AS-protein complexes from formaldehyde crosslinked macrophage cellular extracts, according to a modified Chromatin Isolation by RNA Purification (CHIRP) method.22 We separated lncLrrc55-AS-associated proteins by gel electrophoresis and performed mass spectrometry (MS) (Supplementary information, Fig. S7a, Table S3). Among the candidates, we identified the protein phosphatase methylesterase 1 (PME-1) in a band within the 40–55 kD range that specifically co-purified with lncLrrc55-AS from SeV-infected macrophages (Fig. 5a).
Identification of protein phosphatase methylesterase PME-1 as a binding partner of lncLrrc55-AS. a RT-qPCR analysis of the RNA retrieval ratio of lncLrrc55-AS purified by hybridizing with antisense lncLrrc55-AS probes from whole cell extracts of peritoneal macrophages infected with SeV for 12 h (left). SDS-PAGE with a silver nitrate staining analysis of proteins co-purified with lncLrrc55-AS (right). The arrow indicates protein bands that were subjected to mass spectrometry analysis. NC, no-targeting probe control. b, c RNA pull-down experiments using in vitro transcribed biotinylated lncLrrc55-AS or negative strand RNA (NS RNA) as a control to retrieve PME-1 from HEK293T cell extracts (b) or recombinant His-tagged PME-1 protein collections (c). RNA retrieval ratio of the RNA pull-down assay was assessed by RT-qPCR (b left). PME-1 retrieval by the RNA pull-down assay was assessed by western blot (b right and c). d RNA immunoprecipitation (RIP) experiments were performed with anti-PME-1 antibody in macrophages infected with SeV for 12 h. Immunoprecipitation of PME-1 was assessed by western blot. Co-purified RNAs were assessed by RT-qPCR analysis. e RIP experiments were performed with anti-PME-1 antibody in NIH/3T3 cells infected with VSV for 10 h. Immunoprecipitation of PME-1 and co-immunoprecipitation of PP2A-C were assessed by western blot. Co-purified RNA was assessed by RT-qPCR analysis. f RNA FISH-immunofluorescence detection of endogenous lncLrrc55-AS molecules and PME-1 in macrophages infected with SeV for 12 h. Scale bar, 10 µm. g RIP experiments were performed with anti-Flag antibody-coupled beads in NIH/3T3 cells transfected with full-length or truncated PME-1-Flag plasmids. LncLrrc55-AS retrieval was assessed by RT-qPCR. Schematic of PME-1 and truncated PME-1 used in the study (left). ΔC-arm, carboxyl-terminal arm (residues 371–385) truncated; ΔN-arm, amino-terminal arm (residues 1–64) truncated; ΔC&N-arm, residues 1–64 and 371–385 truncated; ΔCD, cap domain (residues 191–301) deleted; Δ107–113 aa, residues 107–113 deleted. Data are from three independent experiments (a, b left, d left, e, g means ± SEM) or are representative of three independent experiments with similar results (a, b, d right, c, e left, f). *P < 0.05, ***P < 0.005, ND, not detected (Student’s t-test or ANOVA)
RNA pull-down analyses confirmed that lncLrrc55-AS bound to PME-1 in 293T cell extracts and to His-tagged PME-1 recombinant protein collections in vitro (Fig. 5b, c). RNA immunoprecipitation (RIP) revealed that lncLrrc55-AS co-immunopurified with endogenous PME-1 from peritoneal macrophages infected with SeV (Fig. 5d), and with ectopically expressed PME-1-Flag from NIH/3T3 cells infected with VSV (Fig. 5e). Moreover, using an RNA FISH-immunofluorescence assay, we found that lncLrrc55-AS co-localized with PME-1 in the cytoplasm of SeV-infected macrophages (Fig. 5f). Previous studies have detected PME-1 mainly in the nucleus of HeLa cells,23 however, our immunofluorescence assay and biochemical analyses detected PME-1 mainly within the cytoplasm of peritoneal macrophages (Fig. 5f; Supplementary information, Fig. S7c).
To map the lncLrrc55-AS-interacting region within PME-1, we performed RIP with a series of Flag-tagged PME-1 truncations (Fig. 5g; Supplementary information, Fig. S7b). We found that both the amino (N-) and carboxyl (C-) termini of PME-1 were necessary for interaction with lncLrrc55-AS (Fig. 5g). Previous structural analyses reported that the α4, α5, α6, α7 helices of human PME-1 form a cap domain above the active pocket domain,24 but we found that the cap domain was not required for interaction with lncLrrc55-AS. Our MS identification of the PME-1/lncLrrc55-AS interaction suggested that 7 residues (residues 107–113) within the α/β hydrolase fold domain of the PME-1 N-terminus might be important for binding to lncLrrc55-AS. To test this hypothesis, we evaluated a PME-1 mutant lacking residues 107–113. As hypothesized, RIP revealed that this mutant did not interact with lncLrrc55-AS (Fig. 5g). These data suggest that multiple regions within PME-1 are involved in its interaction with lncLrrc55-AS.
To investigate whether PME-1, like lncLrrc55-AS, affects virus-induced IFN-I production, we silenced PME-1 in mouse macrophages (Fig. 6a). We found that silencing of PME-1 reduced the expression of Ifna4 and Ifnb mRNAs in macrophages infected with VSV or SeV (Fig. 6a; Supplementary information Fig. S8a), similar to lncLrrc55-AS silencing (Fig. 2d; Supplementary information, Fig. S4d). To further validate our findings, we utilized CRISPR-Cas9 technology to knock out PME-1 in the macrophage cell line RAW264.7 (Supplementary information, Fig. S8b, c). We found that PME-1 KO RAW264.7 cells displayed a decreased expression of Ifna4 and Ifnb mRNAs upon infection with VSV (Fig. 6b). Furthermore, the VSV titer 12 h after VSV infection was higher in culture supernatant from PME-1 KO RAW264.7 cells than from WT cells (Fig. 6c). Phosphorylation of IRF3 after infection with VSV or SeV was also reduced in PME-1-deficient macrophages compared to controls (Fig. 6d; Supplementary information, Fig. S8d). Thus, PME-1 promotes IRF3-mediated IFN-I production in virus-infected macrophages, similar to lncLrrc55-AS.
LncLrrc55-AS inhibits PP2A-mediated negative regulation of IRF3-induced IFN-I production via PME-1. a RT-qPCR analysis of Ifna4 and Ifnb mRNA expression in non-target siRNA control (NC)- or PME-1-silenced peritoneal macrophages infected with VSV for 12 h. The silencing efficiency of three siRNAs targeting PME-1 was confirmed by western blot (right). b RT-qPCR analysis of Ifna4 and Ifnb mRNA expression in WT or PME-1 KO RAW264.7 cells infected with VSV for 12 h. c Determination of virus loads by TCID50 assay in the supernatant from VSV-infected WT or PME-1 KO RAW264.7 cells for 12 h. d Western blot analysis of phosphorylated (p-) or total proteins in lysates of WT and PME-1 KO RAW264.7 cells infected with VSV for the indicated hours. e RT-qPCR analysis of Ifna4 and Ifnb mRNA expression in NC- and PME-1-, lncLrrc55-AS- or double-silenced peritoneal macrophages infected with VSV for 12 h. f Western blot analysis of p-IRF3 or total IRF3 proteins in lysates of NC- or lncLrrc55-AS-silenced WT and PME-1 KO RAW264.7 cells infected with VSV for the indicated hours. g RT-qPCR analysis of Ifna4 and Ifnb mRNA expression in NC- and PP2A-C-, lncLrrc55-AS- or double-silenced peritoneal macrophages infected with VSV for 12 h. h RT-qPCR analysis of Ifna4 and Ifnb mRNA expression in NC- or lncLrrc55-AS-silenced macrophages pre-treated with Okadaol (50 nM) or DMSO 1 h before infection with VSV for 12 h. i Determination of virus loads by TCID50 in the supernatant from NC- or lncLrrc55-AS-silenced macrophages pre-treated with Okadaol (50 nM) or DMSO 1 h before infection with VSV for 12 h. Data are from three independent experiments (a–c, e, g–i, means ± SEM) or are representative of three independent experiments with similar results (d, f). *P < 0.05, **P < 0.01, ***P < 0.005; ns, no significance (Student’s t-test or ANOVA)
To clarify how lncLrrc55-AS and PME-1 promote IRF3 signaling and IFN-I production, we examined the effects of silencing both lncLrrc55-AS and PME-1 in macrophages. We found that silencing of lncLrrc55-AS reduced IFN-I production upon VSV or SeV infection of control macrophages, but not PME-1-silenced macrophages (Fig. 6e; Supplementary information, Fig. S8e). In addition, silencing of lncLrrc55-AS in PME-1 KO RAW264.7 cells did not further reduce VSV-induced phosphorylation of IRF3 (Fig. 6f). Thus, PME-1 is required for lncLrrc55-AS to promote IRF3 signaling and IFN-I production after viral infection.
LncLrrc55-AS promotes IRF3 signaling by blocking the inhibitory activity of protein phosphatase PP2A
PME-1 is a protein phosphatase methylesterase that inactivates PP2A, a critical negative regulator of IRF3 signaling and thus IFN-I production.7,23,24,25,26 To verify that PP2A negatively regulates IRF3 signaling and thus IFN-I production, we silenced the PP2A catalytic subunit (PP2A-C) in mouse macrophages. Silencing of PP2A-C enhanced IRF3 phosphorylation in SeV-infected macrophages (Supplementary information, Fig. S9a, b). In addition, we found that the treatment with Okadaol, an inhibitor of PP2A activity, enhanced IRF3 phosphorylation in VSV-infected macrophages (Supplementary information, Fig. S9c, d), in accordance with previous reports.7,27 Although PP2A has been reported to target STAT1, p38 and the ERK signaling pathway,28,29 here we did not detect the change of phosphorylation of STAT1, p38 and ERK when PP2A was silenced (Supplementary information, Fig. S9b). Furthermore, silencing of PP2A or Okadaol-mediated inhibition of PP2A promoted IFN-I production in virus-infected macrophages, and abrogated the effects of lncLrrc55-AS silencing (Fig. 6g, h; Supplementary information, Fig. S9e). In addition, silencing of lncLrrc55-AS in PP2A-inhibited macrophages did not lead to an increased VSV titer in cell supernatants (Fig. 6i). Together, these results suggest that lncLrrc55-AS promotes antiviral IFN-I production by inhibiting PP2A-mediated negative regulation of IRF3 signaling.
LncLrrc55-AS strengthens IRF3 signaling by promoting PME-1-mediated demethylation and deactivation of PP2A
PME-1 has been shown to deactivate the phosphatase activity of PP2A by demethylating leucine 309 within PP2A-C.24 We hypothesized that the lncLrrc55-AS-PME-1 complex promotes IFN-I production by PME-1-mediated deactivation of PP2A. Indeed, we found that disruption of lncLrrc55-AS enhanced the phosphatase activity of PP2A in virus-infected macrophages, similar to silencing of PME-1 (Fig. 7a, b). Interestingly, PP2A phosphatase activity was only slightly affected by viral infection in control macrophages (Fig. 7a, b), suggesting that the virus-induced upregulation of lncLrrc55-AS may restrain PP2A activity in macrophages.
LncLrrc55-AS promotes the deactivation and demethylation of PP2A. a Phosphatase activity analysis of PP2A in lysates from non-target siRNA control (NC)- or PME-1-silenced macrophages infected with SeV for 12 h. b Phosphatase activity analysis of PP2A in lysates of NC- or lncLrrc55-AS-silenced macrophages infected with SeV for 12 h (left), and in WT or lncLrrc55-AS−/− macrophages infected with VSV for 12 h (right). c, d Western blot analysis of demethylated PP2A-C (dmPP2A-C) or total PP2A-C proteins in lysates of NC- and PME-1- or lncLrrc55-AS-silenced peritoneal macrophages with SeV infection for the indicated hours. e, f Western blot analysis of dmPP2A-C or total PP2A-C proteins in lysates from WT and lncLrrc55-AS KO NIH/3T3 cells (e), and from EV Ctrl- or lncLrrc55-AS-transfected lncLrrc55-AS KO NIH/3T3 cells (f) infected with VSV for the indicated hours. g, h Demethylation level by western blot analysis (g) and phosphatase activity assay (h) of recombinant PP2A-C proteins catalyzed by PME-1 in vitro, with increasing amount of lncLrrc55-AS added to the catalytic reaction system. The nlncLrrc55-AS: nPME-1 indicates the molar ratio of lncLrrc55-AS to PME-1. Data are from three independent experiments (a, b, h means ± SEM) or are representative of three independent experiments with similar results (c–g). **P < 0.01, ***P < 0.005 (Student’s t-test)
Next, we investigated whether lncLrrc55-AS affects PME-1-mediated demethylation of PP2A-C. We found that silencing of PME-1 inhibited the demethylation of PP2A-C in SeV-infected macrophages, as expected (Fig. 7c). Importantly, deficiency of lncLrrc55-AS also inhibited PP2A-C demethylation in SeV-infected macrophages or VSV-infected NIH/3T3 cells (Fig. 7d, e). In addition, ectopic expression of lncLrrc55-AS rescued the demethylation of PP2A-C in lncLrrc55-AS-KO NIH/3T3 cells infected with VSV (Fig. 7f). At last, we confirmed that lncLrrc55-AS directly promoted PME-1-mediated demethylation of PP2A in vitro and reduced the in vitro phosphatase activity of PP2A (Fig. 7g, h; Supplementary information, Fig. S9f).
Although we did not detect an interaction between PP2A and lncLrrc55-AS via RNA pull-down or RIP, we found that both lncLrrc55-AS and PP2A co-immunoprecipitated with PME-1 (Fig. 5e). In addition, lncLrrc55-AS, PME-1 and PP2A-C co-localized in SeV-infected macrophages, as assayed by RNA FISH- immunofluorescence (Fig. 8a). To investigate how lncLrrc55-AS promotes PME-1-mediated demethylation and deactivation of PP2A, we evaluated the PME-1/PP2A-C interaction and observed an increase of the colocalization of PME-1 and PP2A in macrophages infected with SeV, an effect not observed in non-infected cells, nor in lncLrrc55-AS-deficient macrophages infected with SeV (Supplementary information, Fig. S10a; Fig. 8b). By immunoprecipitation, we found that this interaction was diminished in lncLrrc55-AS-deficient macrophages or RNA-digested macrophages infected with VSV (Fig. 8c, d). Furthermore, ectopic expression of lncLrrc55-AS promoted the interaction between PME-1 and PP2A-C in HEK 293T cells (Fig. 8e). Thus, lncLrrc55-AS promotes the interaction between PME-1 and PP2A-C, which drives PME-1-mediated demethylation and deactivation of PP2A (Supplementary information, Fig. S10b).
LncLrrc55-AS promotes the interaction between PME-1 and PP2A. a RNA FISH-immunofluorescence detection of endogenous lncLrrc55-AS, PME-1, and PP2A-C in peritoneal macrophages from WT or lncLrrc55-AS−/− non-infected mice and mice infected with SeV for 12 h. Scale bars, 10 µm. b Quantitative analysis of colocalization rate for PME-1 and PP2A in NC- or lncLrrc55-AS-silenced peritoneal macrophages after SeV infection for 12 h. c Interaction between PME-1 and PP2A in RAW264.1 cells. NC- or lncLrrc55-AS-silenced cells were infected with VSV for the indicated hours, followed by immunoprecipitation with anti-PME-1 antibody. The immunoprecipitated PME-1 and PP2A were detected by western blot. d Interaction between PME-1 and PP2A in WT and lncLrrc55-AS−/− peritoneal macrophages infected with SeV for the indicated hours. Samples isolated from cells were equally divided into three parts. All parts were digested with Rnase A or Rnase T1 (10 U/mL), followed by immunoprecipitation with anti-PME-1 antibody (two parts) or with normal IgG antibody (one part; control). The immunoprecipitated PME-1 and PP2A were detected by western blot. e Interactions of PME-1 and PP2A with lncLrrc55-AS in an overexpression system. HEK293T cells were transfected with plasmids encoding PME-1-Flag, PP2A-C-Myc, and lncLrrc55-AS, followed by immunoprecipitation with anti-Flag antibody-coupled beads. The immunoprecipitated PME-1-Flag and PP2A-C-Myc were detected by western blot analysis. HEK293T cells transfected with empty vector plasmids were used as controls. Data are from three independent experiments (b, means ± SEM) or are representative of three independent experiments with similar results (a, c–e). *P < 0.05 (Student’s t-test)
Discussion
Our work identified a new lncRNA, lncLrrc55-AS, that enhances IRF3 signaling and virus-induced IFN-I production by promoting PME-1-mediated deactivation of PP2A, a key inhibitor of IRF3 phosphorylation. The expression of lncRNAs can be regulated by various factors, including hormones, stress signals, cytokines, homeostatic signals, and DNA damage and repair signals.30,31,32 Like traditional interferon-stimulated genes (ISG) whose transcriptional output increases in response to IFN, several lncRNAs are induced by IFN and regulate the innate response or inflammation.33,34 We found that lncLrrc55-AS can be induced by viral infection or innate stimuli via the IFN-JAK-STAT signaling pathway. Our findings imply that the expression of lncLrrc55-AS is in accordance with a classical ISG induction, but to obtain molecular insight into the transcriptional regulation of this lncRNA requires further study.
LncRNAs can regulate immune responses and inflammation epigenetically by binding to chromatin modifiers, including hnRNPL, hnRNP-A/B and hnRNP-A2/B1, or transcription-related factors, such as SFPQ.13,35,36,37 In addition, lncRNAs can regulate immune responses non-epigenetically by affecting protein activity or protein-protein interactions.38,39,40 By using several approaches widely used in RNA-protein binding study, in this study we have demonstrated that lncLrrc55-AS can strengthen IRF3 phosphorylation through its specific interaction with PME-1.
IFN production is thought to be regulated mostly by PTMs of upstream components in the signaling pathway, including sensors, adaptors, and signal transport molecules.4,5,41 IRF3 can be deactivated by dephosphorylation or ubiquitination,7,42 but how IRF3 activation levels are maintained to ensure IFN-I production is poorly understood. Research into the IRF3 regulation of IFN-I production has focused on the regulation of upstream molecular signals, including epigenetic mechanisms such as m6A modification of Mavs, Traf3 and Traf6.43 We found that lncLrrc55-AS enhances IRF3 phosphorylation by promoting PME-1-mediated deactivation of its inhibitor PP2A. Our findings underscore an important role for cytoplasmic lncRNAs in the regulation of IRF3 PTMs via RNA-protein interactions. Other cytoplasmic lncRNAs have been shown to regulate inflammation by forming complexes with transcription factors such as NF-κB.40
LncRNAs typically interact with proteins to exert their functions.44 Canonical RNA binding proteins (RBPs) usually possess RNA-binding domains (RBDs) such as the RNA-recognition motif, K-homology domain, double-stranded RBD, S1 domain, and zinc finger PAZ and PIWI domains.45,46 However, some proteins without canonical RBDs interact directly with RNA.17 Although our structure prediction software did not identify canonical RBDs within PME-1, our data strongly suggest that PME-1 directly binds lncLrrc55-AS. LncRNA can regulate the function of protein by altering the subcellular location or structure, and protein-protein interactions or complex assembly by directly interacting with a specific protein.13,15,31,47 Although our results indicate that lncLrrc55-AS does not affect the expression or the cytoplasmic/nuclear distribution of PME-1 in macrophages upon viral infection, we propose that lncLrrc55-AS mediates the PME-1-PP2A interaction via regulation of PME-1 structural conformation or biological function, but the precise details require further structural and biochemical analysis.
An effective innate immune response to viral infection is dependent on broad innate programs, and is regulated by complex mechanisms to eliminate invading pathogens but avoid damage to the host.48 Our results suggest that infection-inducible lncRNAs can positively regulate protein-protein interactions to enhance the innate immune response. Thus, manipulating lncRNA levels and function may be a useful strategy to explore mechanisms of infection and immunity, and potentially to control autoimmune disease or chronic inflammation.
Materials and methods
Mice
C57BL/6 mice were obtained from Joint Ventures Sipper BK Experimental Animals (Shanghai, China). Ifnar1−/− mice were obtained from Jackson Laboratories. Mice were kept and bred in SPF grade conditions. Irf3−/− mice were kindly provided by Dr. Tadatsugu Taniguchi (the Graduate School of Medicine and Faculty of Medicine, University of Tokyo). All animal experiments were conducted according to the National Institutes of Health Guide for Care and Use of Laboratory Animals with approval of the Scientific Investigation Board of Second Military Medical University, Shanghai, China.
lncLrrc55-AS KO mice were generated by replacing the first exon of lncLrrc55-AS with a GFP cassette followed by a poly A signal under the control of the endogenous promoter of lncLrrc55-AS. Specifically, the first exon of lncLrrc55-AS was knocked out using CRISPR-Cas9 system. A fragment of a GFP cassette followed by a poly A signal was knocked into the homologous locus by homologous recombination. Plasmids expressing guide RNA, Cas9 protein, and the donor plasmid containing the insertion sequence were co-microinjected into C57BL/6 mouse embryonic stem (ES) cells, followed by selection with PCR (Supplementary information, Fig. S4e, f). Positive ES cells were injected into blastocytes to generate chimeric mice. Animals were reproduced at expected Mendelian frequencies with no gender bias. The sgRNA targeting sequences were as follows: EA-F, 5′-TAG GCT CTC TGC AAT GGC TGA C-3′, coupled with EA-R, 5′-AAA CGT CAG CCA TTG CAG AGA G-3′; EB-F, 5′-TAG GGG TAA GCA GGT TGC TGA G-3′, coupled with EB-R, 5′-AAA CCT CAG CAA CCT GCT TAC C-3′.
Cells
RAW264.7, NIH/3T3, HEK293T, MDCK, MLE12, 3LL and Hepa cells were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco’s Modified Eagle’s medium (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL). Mouse peritoneal macrophages were prepared as described.20 Cells were seeded into cell culture plates and incubated to reach a density of 70% for further treatments.
The knockout strategy for lncLrrc55-AS in NIH/3T3 cells was the same as that in mice. The knockout of pme1 in RAW264.7 cells was constructed by interrupting the open reading frame (ORF) of pme1 to break the translation of mRNAs using CRISPR-Cas9 system (Supplementary information, Fig. S7b). The target site for pme1 was 5′-TTG TGG CTC TGG ATC TGCG-3′ (in exon #4).
Reagents
Antibodies against Myc (9E10), Flag tag (H-70), GAPDH, p-IRF3 (4947), IRF3 (4302), TBK1 (3013), p-TBK1 (5483), p-p65 (3033), p65 (8242), p38 (8690), p-p38 (4511), ERK (4695), p-ERK (4370), STAT1 (14994) and p-STAT1 (8826) were from Cell Signaling Technology. Anti-PME-1 antibody was from Santa Cruz Biotechnology (sc-55472). Anti-PP2A-C antibody was from Abcam (ab32104). Anti-demethylated PP2A-C antibody was from Millipore (05–577). Protein G magnetic beads were from Cell Signaling Technology (9006). Anti-flag-M2 magnetic beads were from Sigma. Streptavidin C1 beads were from Thermo Fisher. ELISA kits for mouse IFNα and IFNβ were from PBL Biomedical Laboratories and for mouse IL-6 and TNFα were from R&D Systems.
Virus
VSV (Indiana Strain) and VSV-GFP were propagated and amplified by infection of a monolayer of HEK293T cells. HSV-1 was from the Cell Resource Center of the Shanghai Academy of Sciences, Chinese Academy of Sciences. SeV (Senda virus) was propagated and amplified by inoculating chick embryo allantoic fluid. The viral titer of VSV was determined by TCID50 on HEK293T cells, and the viral titer of HSV-1 was determined using MDCK cells as previously described.20 The viral titer of SeV was determined by direct hemagglutination (HA) assay. Cells were infected with VSV (MOI = 1), SeV (MOI = 1), or HSV-1 (MOI = 10) for the indicated hours. Influenza virus A methods (Puerto Rico/8/1981H1N1(PR8)) were described previously.20
Mouse experiments
Sex- and age-matched littermates were infected with VSV (5 × 107 pfu/g) by intraperitoneal injection for in vivo experiments. For survival experiments, mice were intraperitoneally injected with 1 × 108 pfu/g VSV. Blood samples were collected from the orbital vein, and tissue samples were collected immediately after mice were sacrificed by cervical dislocation.
RACE and cloning of full-length lncLrrc55-AS
The 3′ and 5′ RACE was performed using the SMARTer RACE 5′/3′ kit (Clontech) following the manufacturer’s instructions. RNA was extracted from mouse peritoneal macrophages. Primers used for 3′ and 5′ RACE were as follows and designed based on known sequence information:
CAGGGACAGATGGAGCCTCACTCATTGGGATGAG (3′ specific); CCAATGAGTGAGGCTCCATCTGTCCCTGAGCAAG (5′ specific).
Plasmid- and lentivirus-mediated ectopic expression
cDNA encoding mouse PME-1 (ENSMUSG00000030718) or PP2A-C (ENSMUSG00000020349) was amplified from mouse peritoneal macrophages and cloned into pcDNA3.1-Flag or pcDNA3.1-Myc eukaryotic expression vectors, respectively. Deleted, truncated mutants were generated by PCR-based amplification with the construct encoding WT protein as the template. Each construct was confirmed by sequencing. Plasmids were transiently transfected into cells with Jetprime reagents (Polyplus Transfection) according to the manufacturer’s instructions.
cDNA of lncLrrc55-AS was cloned into the lentivirus expression vector pCDH-CMV-MCS-EF1-Puro, and packaged into lentivirus particles. Macrophages were infected with polybrene (Polyplus Transfection) according to the manufacturer’s instructions. Lentivirus containing empty vector was used as a negative control. Infection efficiency was determined by GFP fluorescence and qPCR analysis of total RNA from infected cells. VSV or SeV was added after 36 h.
Small RNA-mediated interference
Small interference RNAs (siRNAs) were transfected into the indicated cells using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher) per standard procedures. The specific siRNA target sites for lncLrrc55-AS were #1 (GGA GAAGACTCTTGCTCAG) and #2 (CCTGTAGTTTGGTTATTCT). Target sites for PME-1 were #1 (GCAGCTAACCTGGTACCA), #2 (CCGCCATGGATGCACTTA) and #3 (CCATAGTGGAAGGAATCA). The target site for PP2A-C was GACGAGTGTTTAAGGAAAT. A non-targeting siRNA was used as a control.
Cell fraction isolation
Cells were harvested from culture plates using a rubber cell scraper and washed twice with cold PBS. Cells were then suspended in PBS containing 0.1% NP-40, and gently crashed. Half of the suspended cells were removed for lysis to serve as whole cell controls. The remaining half of the cells was centrifuged at 8000 rpm, 4 °C. The supernatant was removed to isolate cytoplasm. The precipitate was washed with 0.05% NP-40, and removed to isolate nucleus. The cell fractions were each divided into two parts for RNA isolation and protein analysis. The expression of target genes in individual fractions was normalized to their expression level in the input RNA, which was set to 100%.
RNA extraction and RT-qPCR
Total RNA was purified from cells and tissues using either TRIzol reagent (Ambion) or an RNeasy RNA extraction kit (Qiagen), respectively, per the manufacturers’ instructions. Genomic or plasmid DNA was eliminated by treatment with DNase1 (Sigma). Equal amounts of RNA were reverse-transcribed using an iScript cDNA synthesis kit (ToYoBo). Diluted cDNAs were subjected to qPCR analysis using SYBR Green Supermix reagent (ToYoBo) according to the manufacturer’s instructions, followed by melting curve analysis. RT-qPCR primer sequences are provided in Supplemental information, Table S1. Gene expression levels were normalized to the housekeeping hypoxanthine guanine phosphoribosyl transferase (HPRT) gene.
Dual-luciferase reporter assays
Dual-luciferase reporter assays were performed as previously described.7 Briefly, 293T cells were seeded on 24-well plates and transfected the following day with pIFNβ-luc, pRL-TK (constitutively expressing Renilla luciferase), and the described gene constructs. Cells were analyzed with a Dual-Luciferase reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase activity was normalized to pRL-TK signal.
RNA FISH-immunofluorescence microscopy
Fluorescein isothiocyanate (FITC)-conjugated lncLrrc55-AS probes were generated by Biosearch Technologies. Sequences are listed in Supplementary information, Table S1. LncLrrc55-AS was hybridized with DNA probe sets. PME-1 and PP2A were immunostained with the appropriate antibodies and separately immunostained with Alexa Fluor 594-conjugated anti-mouse IgG and Cy5-conjugated anti-rabbit IgG, respectively. Images were obtained with an Olympus FV1000 laser-scanning confocal microscope.
Modified chromatin isolation by RNA purification
CHIRP-q-PCR and CHIRP-MASS assays were performed as previously described.22 Briefly, 5′ biotin-labeled antisense DNA probes for lncLrrc55-AS were designed by Biosearch Probe Designer. Sequences are listed in Supplementary information, Table S1. Probe pools were hybridized with whole cell extracts from ~3 × 107 macrophages. Hybridization components were incubated with Streptavidin-magnetic C1 beads. Pull-down components were divided into two parts: one for RNA purification efficiency, and the other separated by SDS-PAGE and silver staining. Differential bands enriched by lncLrrc55-AS were analyzed by LTQ Orbitrap XL mass spectrometry.
In vitro transcription of RNA
Biotinylated lncLrrc55-AS RNA, with the antisense strand for lncLrrc55-AS as control, were obtained with T7 RNA Polymerase (NEB) and labeled with biotin RNA labeling mix (Roche) in vitro. The transcribed RNAs were treated with on-column DNase I digestion (Qiagen) to eliminate the template DNA, and purified using an RNeasy Mini Kit (Qiagen). Biotinylated RNA was allowed to form proper secondary structures as follows: RNA was heated to 90 °C for 2 min, put on ice for 2 min, supplied with RNA structure buffer (10 mM Tris, pH 7, 0.1 M KCl, 10 mM MgCl2), and then shifted to room temperature for 20 min.
RNA pull-down assay
RNA pull-down assays were conducted as previously described.20 Briefly, each binding reaction was performed using 1 µg of biotinylated lncLrrc55-AS and antisense strand control RNAs, and incubated with whole cell extracts separated from 1 × 107 293T cells. Pull-down assays of biotinylated RNAs with His-tagged PME-1 recombinant protein collections in vitro were performed as above. Briefly, recombinant His-tagged PME-1 was expressed and purified using a prokaryotic expression system. One microgram biotinylated RNA was incubated with different amounts of His-tagged PME-1 recombinant protein (0.1, 0.5 and 1 µg) in binding buffer at 37 °C for 1 h. Biotin-RNA/protein complexes were captured using 30 µL Streptavidin C1 beads (Invitrogen) in RIP buffer. Pull-down components were eluted by heating samples at 95 °C for 5 min in 2× Laemmli buffer containing 1% SDS, and separated by SDS-PAGE. Differential bands enriched by lncLrrc55-AS were analyzed by immunoblotting.
RNA immunoprecipitation
Peritoneal macrophages were dissolved with RNase-free cell lysis buffer. Samples were sonicated on ice three times and centrifuged at 12,000× g for 10 min. Supernatants were diluted in 1 mL fresh RIP buffer and precleared with Protein G beads following overnight incubation with specific antibodies at 4 °C. Protein G beads were then added and incubated at 4 °C for 2 h. Total RNA or protein was extracted from the eluent. Immunoprecipitation efficiency was analyzed by immunoblotting and lncLrrc55-AS enrichment was analyzed by RT-qPCR.
Phosphorylation activity
The phosphorylation activity of PP2A was detected via PP2A immunoprecipitation using a phosphatase assay kit (Millipore, #2736534) according to the manufacturer’s protocol. Briefly, ~2 × 106 cells were harvested, and PP2A was immunoprecipitated using anti-PP2A antibody and protein A agarose. The purified PP2A enzyme reacted with a phosphopeptide, and the activity was measured by malachite green phosphatase assay read at 650 nm. The phosphorylation activity of the recombinant PP2A-C (Abcam, 128557) (0.75 mg per reaction) was also detected according to the manufacturer’s protocol.
Demethylation assay
The PME-1 to PP2A demethylation assay was conducted according to previous descriptions.24,49 His-tagged PME-1 and GST-tagged PP2A-C recombinant proteins were mixed into demethylation reaction buffer (HEPES-Potassium hydroxide, pH 7.5, 50 mM; Ammonium iron(II) sulfate, 70 µM; α-Ketoglutaric acid, 1 mM; L-Ascorbic Acid Sodium Salt, 2 mM) in an appropriate molar ratio (n/n), and incubated at 37 °C. The reaction was terminated by moving onto ice for immediate immunoprecipitated phosphatase detection, or boiling with SDS-PAGE loading buffer for immunoblotting. The demethylation of the GST-tagged PP2A-C recombinant protein was measured by anti-demethylated PP2A-C antibody. LncLrrc55-AS was added into the demethylation reaction system described above with a molecular molar ratios of 0.25:1, 0.5:1, 1:1, 2:1, 4:1 and 8:1 to PME-1.
Statistical analysis
The statistical significance of comparisons between two groups was determined with an unpaired Student’s t-test. For comparison of more than 2 groups, one-way ANOVA was used. P values < 0.05 were considered statistically significant. Kaplan–Meier survival curves were generated and analyzed for statistical significance with GraphPad Prism6.0.7.
References
McNab, F. et al. Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015).
Hoffmann, H. H., Schneider, W. M. & Rice, C. M. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 (2015).
Wu, J. & Chen, Z. J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015).
Liu, J., Qian, C. & Cao, X. Post-translational modification control of innate immunity. Immunity 45, 15–30 (2016).
Fitzgerald, K. A. et al. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 (2003).
Long, L. et al. Recruitment of phosphatase PP2A by RACK1 adaptor protein deactivates transcription factor IRF3 and limits type I interferon signaling. Immunity 40, 515–529 (2014).
Shen, Q. et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 554, 123–127 (2018).
Schlums, H. et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Marazzi, I., Greenbaum, B. D., Low, D. H. P. & Guccione, E. Chromatin dependencies in cancer and inflammation. Nat. Rev. Mol. Cell Biol. 19, 245–261 (2018).
Chen, Y. G., Satpathy, A. T. & Chang, H. Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 18, 962–972 (2017).
Atianand, M. K. et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 (2016).
Liu, B. et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat. Immunol. 18, 499–508 (2017).
Morchikh, M. et al. HEXIM1 and NEAT1 Long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell 67, 387–399 (2017).
Nishitsuji, H. et al. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc. Natl Acad. Sci. USA 113, 10388–10393 (2016).
Wang, P., Xu, J., Wang, Y. & Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358, 1051–1055 (2017).
Xie, Q. et al. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J. Virol. 92, e00507–e00518 (2018).
Winterling, C. et al. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 11, 66–75 (2014).
Jiang, M. et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906–919 (2018).
Carnero, E. et al. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep. 17, 1013–1028 (2016).
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E. & Chang, H. Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 (2011).
Longin, S. et al. Spatial control of protein phosphatase 2A (de)methylation. Exp. Cell Res. 314, 68–81 (2008).
Xing, Y. et al. Structural mechanism of demethylation and inactivation of protein phosphatase 2A. Cell 133, 154–163 (2008).
Peng, D. et al. A novel function of F-Box protein FBXO17 in negative regulation of type I IFN signaling by recruiting PP2A for IFN regulatory factor 3 deactivation. J. Immunol. 198, 808–819 (2017).
Kaur, A. & Westermarck, J. Regulation of protein phosphatase 2A (PP2A) tumor suppressor function by PME-1. Biochem. Soc. Trans. 44, 1683–1693 (2016).
Albano, C. et al. The total activity of a mixture of okadaic acid-group compounds can be calculated by those of individual analogues in a phosphoprotein phosphatase 2A assay. Toxicon 53, 631–637 (2009).
Meng, G. et al. Combination treatment with triptolide and hydroxycamptothecin synergistically enhances apoptosis in A549 lung adenocarcinoma cells through PP2A-regulated ERK, p38 MAPKs and Akt signaling pathways. Int. J. Oncol. 46, 1007–1017 (2015).
Ivaska, J., Bosca, L. & Parker, P. J. PKCε is a permissive link in integrin-dependent IFN-γ signalling that facilitates JAK phosphorylation of STAT1. Nat. Cell Biol. 5, 363–369 (2003).
Ulitsky, I. & Bartel, D. P. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013).
Munschauer, M. et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 561, 132–136 (2018).
Satpathy, A. T. & Chang, H. Y. Long noncoding RNA in hematopoiesis and immunity. Immunity 42, 792–804 (2015).
Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014).
Carnero, E. et al. Type I interferon regulates the expression of long non-coding RNAs. Front. Immunol. 5, 548 (2014).
Carpenter, S. et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013).
Imamura, K. et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 53, 393–406 (2014).
Ma, H. et al. The Long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 91, e02250–16 (2017).
Gough, D. J. et al. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36, 166–174 (2012).
Yang, F., Zhang, H., Mei, Y. & Wu, M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell 53, 88–100 (2014).
Rapicavoli, N. A. et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2, e00762 (2013).
Liu, B. et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381 (2015).
Chattopadhyay, S. et al. Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. mBio 4, e00636–00612 (2013).
Yu, Y. & Hayward, G. S. The ubiquitin E3 ligase RAUL negatively regulates type I interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33, 863–877 (2010).
Atianand, M. K., Caffrey, D. R. & Fitzgerald, K. A. Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 35, 177–198 (2017).
Nakagawa, S. & Kageyama, Y. Nuclear lncRNAs as epigenetic regulators-beyond skepticism. Biochim. Biophys. Acta 1839, 215–222 (2014).
Hentze, M. W., Castello, A., Schwarzl, T. & Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018).
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011).
Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16, 35–50 (2016).
Dhayalan, A. et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114–26120 (2010).
Acknowledgements
We thank Dr. Pin Wang for technical assistance. This work is supported by grants from the National Natural Science Foundation of China (81788101 to X.C.), the National Key Research & Development Program of China (2018YFA0507403 to X.C.), and CAMS Innovation Fund for Medical Sciences (2016-12M-1-003 to X.C.).
Author information
Authors and Affiliations
Contributions
X.C. designed and supervised the research; Y.Z., M.L., Y.X., Z.L., W.W. and X.L. performed the experiments; Y.M. and L.Z. generated KO mice; Z.S. provided reagents; X.C. and Y.Z. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhou, Y., Li, M., Xue, Y. et al. Interferon-inducible cytoplasmic lncLrrc55-AS promotes antiviral innate responses by strengthening IRF3 phosphorylation. Cell Res 29, 641–654 (2019). https://doi.org/10.1038/s41422-019-0193-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-019-0193-0